Molecular compatibility of malaria parasites in mosquito vectors
Published in General & Internal Medicine
Malaria's global burden
Malaria continues to be one of humanity's deadliest parasitic diseases, causing over 200 million cases and about 600,000 deaths every year. Plasmodium falciparum is responsible for the majority of this impact, especially in sub-Saharan Africa, with 93% of illness and 95% of deaths attributed to the species. Plasmodium vivax is the second most widespread species globally, and is particularly prevalent across Asia and the Americas.
Malaria transmission dynamics
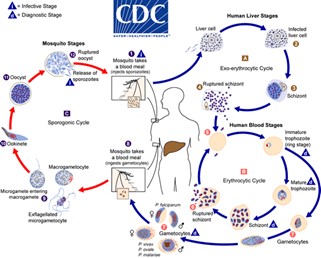
Both P. falciparum and P. vivax are transmitted via bites from more than 70 species of Anopheles mosquitoes worldwide. When a competent mosquito vector takes a blood meal from a human already infected with malaria, the parasite undergoes successive transformations from a gametocyte (taken up in the blood) to oocysts that migrate and invade the mosquito’s midgut. These oocysts then release sporozoites that traverse to the salivary glands, ready to be transmitted to another human host during the mosquito's next blood meal.
Much like in the human-related life cycle stages, the parasites come up against an assortment of immune defences whilst inside their mosquito host, including numerous epithelial, humoral, and cellular components.
Parasites employ a diverse set of molecules to manipulate their host’s sophisticated immune system, as well as variation and adaptation of these molecules as a means of evading immune defences, both in the human and mosquito hosts. In turn, these changes drive adaptive variation in host immune effector molecules that target the parasite, creating an evolutionary arms race between host and parasite.
P47, a key player in mosquito infection and immune evasion
One parasite protein that illustrates this dynamic particularly well is P47, a surface protein expressed during the ookinete stage of the parasite. Pfs47, the P47 protein in P. falciparum, is known help the parasite evade mosquito immune defences by suppressing nitration (an immune reaction in the mosquito midgut epithelium), thereby blocking activation of the complement-like system, a critical component of mosquito immunity.
Researcher have described a “lock-and-key” model, where Pfs47 (key) must match a mosquito midgut receptor named P47Rec (lock) to evade detection. The compatibility of these molecules determines whether the parasite survives or is eliminated by the mosquito immune system.
Pfs47 is highly diverse across parasite populations worldwide, and even single amino acid changes in its second domain (D2) can dictate whether a parasite is compatible with a given mosquito species. This diversity suggests that P. falciparum populations have adapted to local vectors, fine-tuning Pfs47 variants to ensure transmission in different geographic regions.
A similar pattern is seen in P. vivax, whose ortholog (Pv47) is thought to play a comparable role in mosquito immune evasion. Though its mechanisms remain less clear, Pv47 shows strong global structuring, with distinct variants in South America compared with Southeast Asia.
The evolutionary adaptation of Pfs47 and Pv47 to diverse mosquito species not only underscores their importance in parasite–vector interactions but also their potential as transmission-blocking vaccine (TBV) targets. Disrupting the function of these proteins could prevent parasites from evading mosquito immunity, breaking the transmission cycle. Experimental work supports this for Pfs47, while Pv47 is emerging as a promising TBV candidate, though further research is needed to clarify its precise role and its global diversity.
As well laying the foundations of our P47 understanding, research by Alvaro Molina-Cruz and colleagues in the USA has recently investigated the global genetic diversity and evolution of Pv47, and compare it to Pfs47 to further study the potential role of Pv47 in vector compatibility and adaptation of P. vivax parasites to different mosquito species
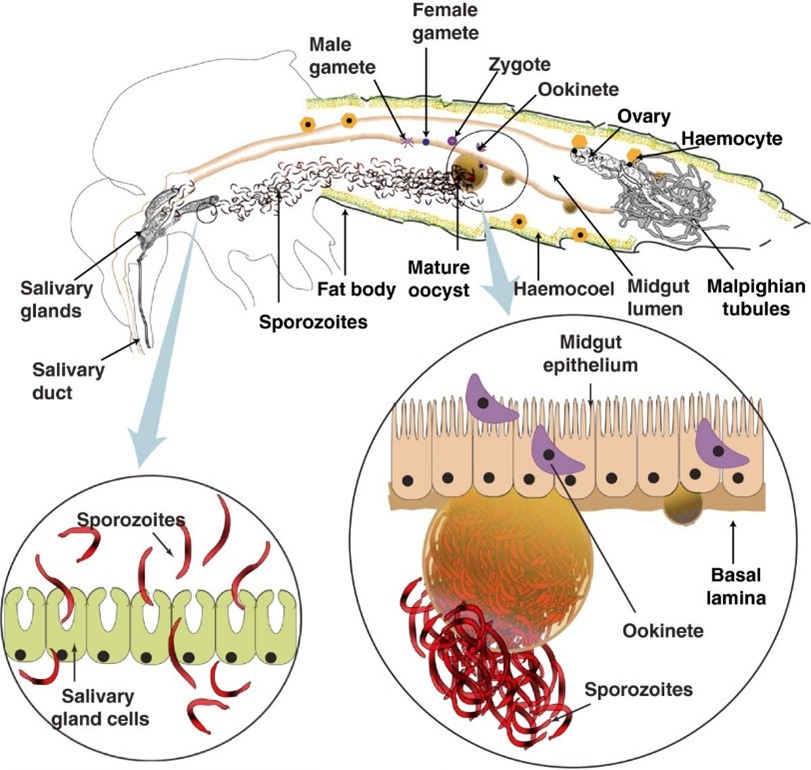
Modified from Sreenivasamurthy et al., 2013. https://doi.org/10.1186/1475-2875-12-216
How the Study Was Done
To understand how Plasmodium vivax and Plasmodium falciparum adapt to different mosquito species around the world, and investigate P47s role, the researchers analysed thousands of Pv47 and Pfs47 gene sequences from P. vivax and P. falciparum, respectively. Together, this made it one of the largest comparisons of these immune-evasion genes to date.
Once the sequences were gathered, the researchers assessed their mutation content, haplotype distribution, genetic diversity between populations, genetic differentiation, and looked for signs of natural selection driving functional changes in the resulting proteins.
By combining global sequencing data, evolutionary analysis, and protein modelling, the researchers could test whether these genes are evolving under pressure from mosquito immune defences.
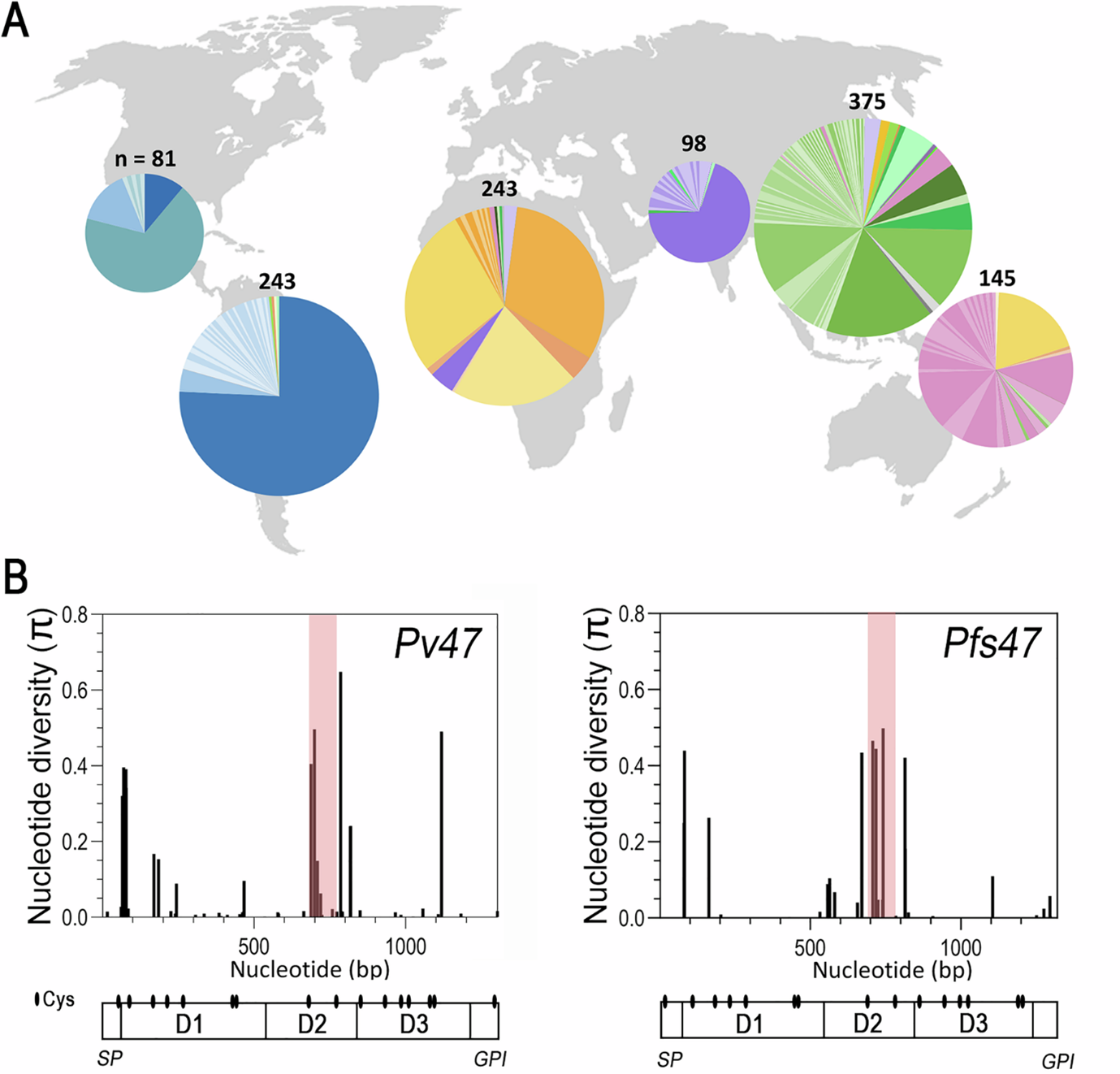
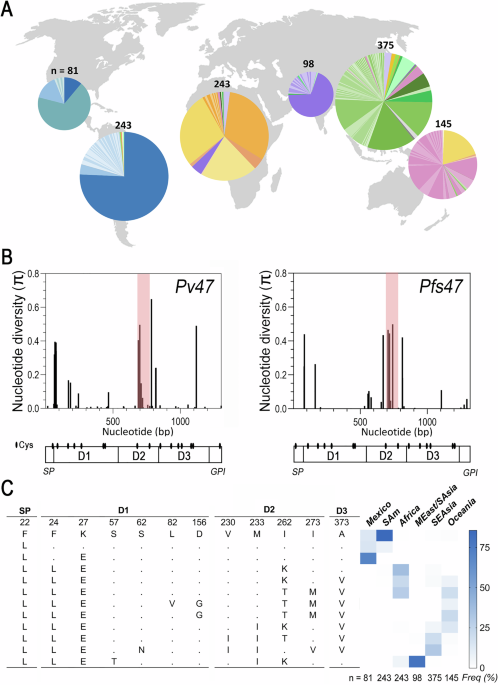
Distribution of Pv47 protein haplotypes globally (A) and Nucleotide diversity for Pv47 and Pfs47 gene coding sequences (B). Modified from Alvaro Molina-Cruz et al., 2025. https://doi.org/10.1038/s41467-025-62680-3
Pv47 is diverse worldwide – and that diversity looks like mosquito-driven selection
The researchers found high genetic diversity among Pv47, with more than 200 unique, often geographically restricted haplotypes discovered, with variants in Asia and Oceania showing the highest levels of diversity. Importantly, and much like observed in P. falciparum, many mutations in the D2 domain of Pv47 showed strong signatures of positive selection.
The fact that both Pv47 and Pfs47 share the same mutational patterns in D2 suggests they serve a similar function helping parasites adapt to local mosquito. The strong regional structuring also makes Pv47 a promising marker of geographic origin in imported malaria cases, just like Pfs47 has already been used.
Strong geographic structuring reveals local adaptation
Pv47 haplotype variants were found to cluster by continent, where, for example, South American parasites were genetically very distinct from those in Africa, Asia, and Oceania-derived parasites. Specific regional mutations stood out, such as F24L (a change from Phenylalanine (F) at position 22 to a Leucine (L)) marking New World haplotypes, and M33I marking African haplotypes.
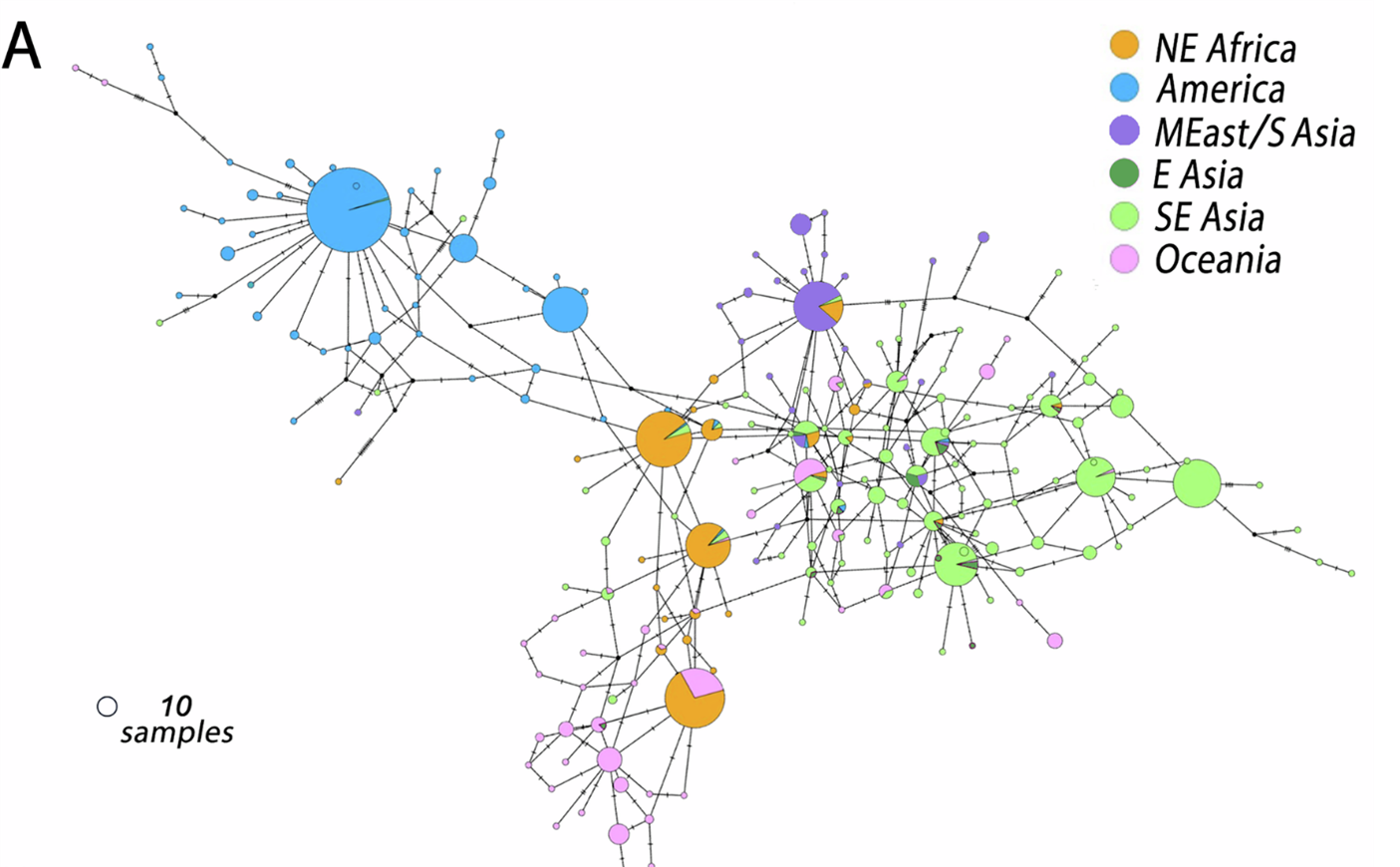
Pv47 haplotype network identified globally. Modified from Alvaro Molina-Cruz et al., 2025. https://doi.org/10.1038/s41467-025-62680-3
South America also showed the lowest genetic diversity, a pattern shared with Pfs47. This likely reflects a combination of history (recent introduction of malaria parasites during European colonisation) and strong selection by local mosquitoes, such as An. albimanus, which are very different from African vectors. In contrast, Oceania (not the parasite’s place of origin) had exceptionally high Pv47 and Pfs47 diversity, possibly reflecting either (a) mosquitoes there are less selective, allowing many haplotypes to coexist, or (b) the region’s very high malaria transmission intensity maintains diverse lineages.
Pfs47 shows parallel patterns, but with some differences
Almost 5,000 Pfs47 sequences from P. falciparum showed similar global structuring and mosquito-driven selection. South American parasites were again genetically distinct, and regional differences were evident within Asia and Africa. Like Pv47, most functional changes clustered in Domain 2.
The parallels between the Pfs47 and Pv47, including similar global structuring and mutation accumulation in the D2 domain, underscore that they are functional analogs shaped by the same evolutionary pressure. Still, Pv47 showed slightly higher diversity overall, consistent with P. vivax’s longer evolutionary history in humans and broader ecological range.
A “molecular switch” in Mexico determines mosquito compatibility
The most striking result came from experimental infections in southern Mexico. Here, two different mosquitoes transmit P. vivax - An. albimanus in lowlands and An. pseudopunctipennis in the foothills.
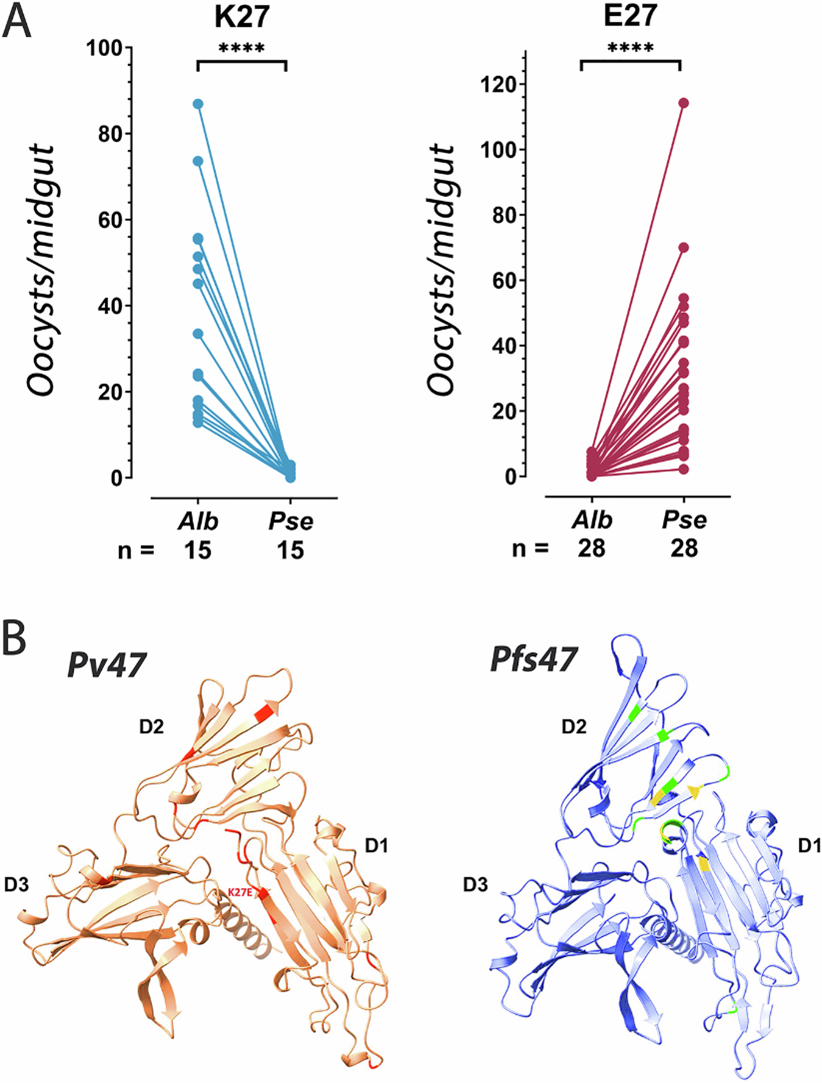
Average number of P. vivax oocysts per midgut in Anopheles albimanus (Alb) and Anopheles pseudopunctipennis (Pse) mosquitoes (A), and predicted Protein structure of Pv47 orthologs (B). From Alvaro Molina-Cruz et al., 2025. https://doi.org/10.1038/s41467-025-62680-3
A single mutation (K27E) on the surface of Pv47 perfectly predicted which mosquito the parasite could infect more efficiently, where K27 parasites infected An. albimanus better and E27 parasites infected An. pseudopunctipennis better.This shows Pv47 can indeed act like a compatibility ‘switch’, determining transmission success in different mosquitoes.
This strengthens the case that Pv47 helps parasites evade mosquito immune defences, just like Pfs47, and suggests Pv47 could be a viable transmission-blocking vaccine target - if antibodies bind these surface regions, they might block parasite development inside mosquitoes.
The Big Picture
Together, these results and interpretations support a unifying hypothesis: Pv47 is a mosquito-compatibility gene, just like Pfs47, and its diversity is driven by adaptation to local vector species worldwide. The gene’s diversity makes it both a marker of parasite origin and a potential weak point for intervention that warrants future investigation. Overall, this valuable work provides new insight into malaria’s evolutionary arms race with mosquitoes and highlights Pv47 as not just a marker of adaptation, but potentially a target for vaccines designed to block transmission.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
-
BugBitten

A blog for the parasitology and vector biology community.
Related Collections
With Collections, you can get published faster and increase your visibility.
Cancer epigenetics and epitranscriptomics
Publishing Model: Open Access
Deadline: Mar 31, 2026
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in