Next generation sequencer repurposed for the oligonucleotide libraries purification
Published in Bioengineering & Biotechnology
Explore the Research
Purification of multiplex oligonucleotide libraries by synthesis and selection - Nature Biotechnology
Accurate oligonucleotide libraries are produced by synthesis and selection.
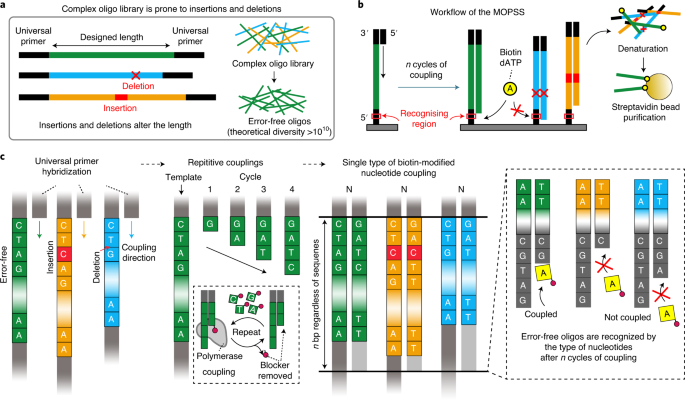
For the last decades, synthetic oligonucleotides (oligos) have bloomed various applications such as nucleic acid-based therapies (CRISPR-Cas9, DNA/RNA vaccines, RNAi), synthetic biology, protein engineering, and DNA-based data storage. Also, nucleic acid-based technologies such as mRNA vaccines or diagnostics are underscored by the COVID-19 pandemic. As they emerge as the most promising weapons against the current pandemic, we expect an increasing need for highly accessible nucleic acid-based technologies. Although oligo synthesis technology has been developing in terms of fidelity and throughput, oligo synthesis is still vulnerable to various synthesis conditions such as humidity, temperature and concentration of reagents. Furthermore, the applications were suffering from the low quality of oligo libraries especially for complex libraries due to a lack of adequate purification method. To solve the problem, our lab was deeply interested in increasing the fidelity of the complex oligo libraries in accordance with our lab’s motto “helping life scientist with technology”.
Six years ago, our lab introduced pyrosequencing-based oligo purification technology [1]. The technology was operated by sequencing the oligo libraries followed by one by one isolation of error-free oligos with a laser-based isolation system. We believe that the technology reduced the cost of gene synthesis by order since the efficiency of gene synthesis largely relies on the fidelity of building blocks that is synthetic oligos. But the throughput which is defined by the number of different oligos that can be simultaneously purified was not suitable to be applied to various state of art nucleic acid-based technologies. Also, we previously utilised a pyrosequencing system (454 Life Sciences) that faded out, and a laser-based isolation instrument, an in-house instrument. The technology was hardly adopted by other researchers from the world. To meet the strong demand for high fidelity complex oligo libraries, we were thirsty for the new technology that can purify complex oligo libraries and that can be immediately adopted by researchers in both academia and industry. To harness these problems, we recently developed multiplex oligonucleotide libraries purification by synthesis and selection (MOPSS) [2]. The project was initiated by simple thinking of “how to measure the length of DNA in a high-throughput manner”. Spacing between nucleotides is 0.34 nm and is too small to physically separate error-free oligos with single-base resolution. Thus, we thought that strategy of counting the number of nucleotides that are coupled from end to end would be better rather than measuring the entire length at once. Then we came up with an idea that maybe we could purify complex oligonucleotide libraries in one pot if we could leverage the fact that most of the errors in synthetic oligo libraries are insertions and deletions (indels). Finally, the idea was refined as following; i) lengths of the oligos in the library is measured by repetitively coupling nucleotides for particular cycles, ii) error-free oligos and oligos with errors are distinguished by the type of nucleotides (i.e. A, G, T, and C) after the specific cycles of couplings, iii) a specific type of biotin-modified nucleotide, for example, biotin-modified dATP is added and is coupled to only error-free oligos followed by selection.
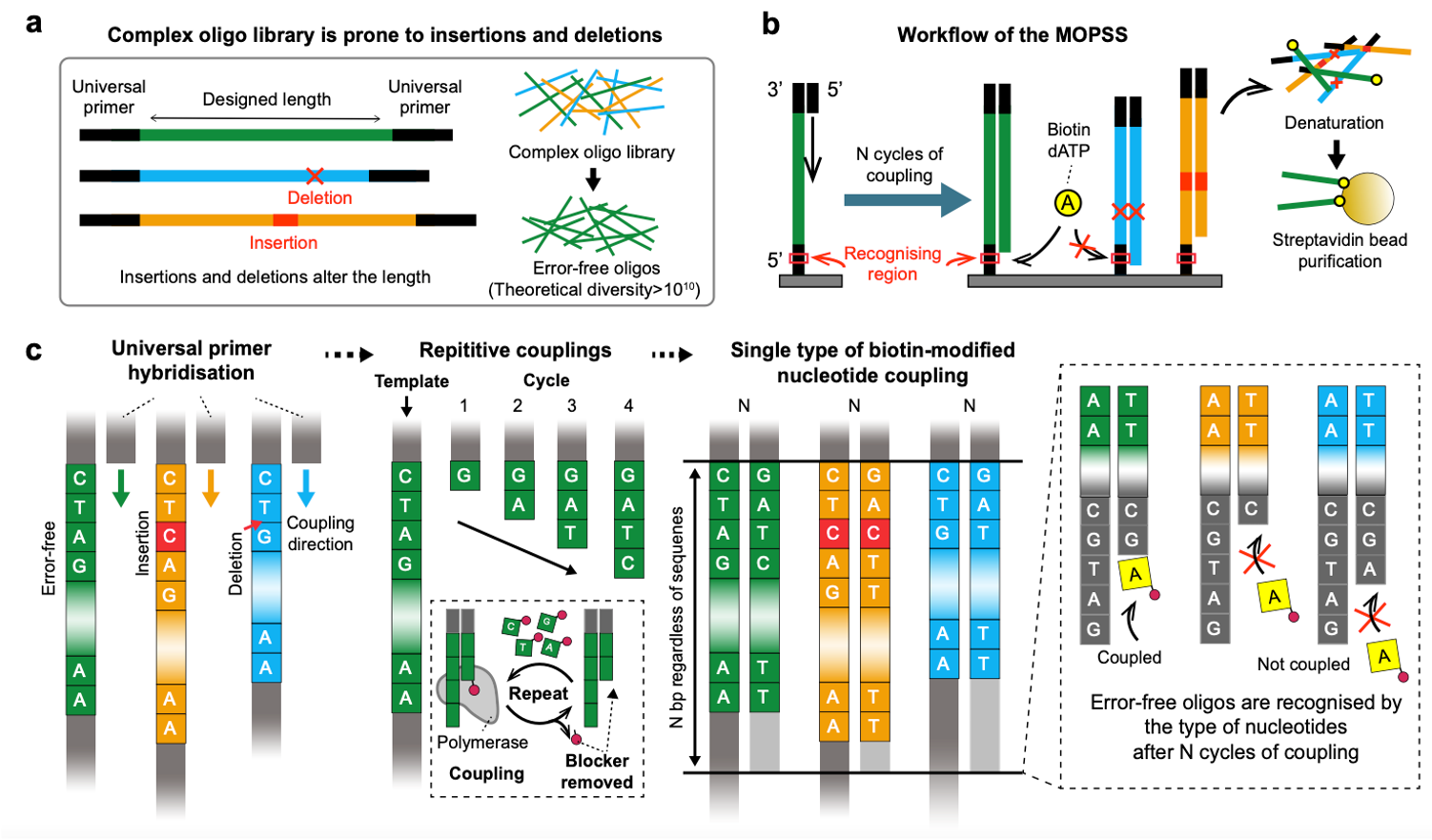
When measuring the lengths of the oligos or repetitively coupling nucleotides, the NGS instrument was used. The main principle behind the NGS instrument is sequencing by synthesis (SBS). SBS is operated by coupling one nucleotide at a time to decipher the sequences of oligos which is identical to our main idea of purification. Depending on the lengths of oligo libraries that we want to purify, the number of coupling cycles must be adjusted. And we hacked the NGS instrument to physically acquire oligos from the flow cell after the intended cycle of couplings or sequencing. Theexperimental design is simple, and any lab with an NGS instrument can adopt this methodology. Since the NGS platform is one of the most widely used and is available to industries, in addition to core and individual labs in academia, we believe that the technology can be easily adopted by researchers.
The MOPSS can be applied to highly complex oligo libraries of different lengths, enabling the simultaneous purification of complex oligo librarieswith different lengths. The proposed technology allows the discarding of oligos with indels by determining the type of nucleotide at the design position; therefore, the purification is unaffected by the oligo sequences, complexity, and differently designed lengths within a library. To harness these advantages, we successfully purified a digital-data-encoded oligo library that was designed with degenerate bases (a combination of two or more nucleotides at a position) and complementarity-determining region (CDR) H3 region encoding oligo libraries with multiple lengths (diversity > 10^9, empirically achieved diversity > 10^6).
Since oligo libraries with high fidelity are fundamental building blocks in various applications, we believe that tackling the major bottleneck of synthesis will allow the technology to flourish and meet the strong demands in numerous research fields. In particular, as nucleic acid-based technologies, such as vaccines or diagnostics, emerge as the most promising weapons against the current pandemic, we expect an increasing need for highly accessible nucleic acid purification methodologies.
References
- Lee, H. et al. A high-throughput optomechanical retrieval method for sequence-verified clonal DNA from the NGS platform. Nat. Commun. 6, (2015).
- Choi, H et al. Purification of multiplex oligonucleotide libraries by synthesis and selection, Nat. Biotechnol. (2021).
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in