One by one - Characterizing anti-bacterial antibody repertoires with single-antibody resolution
Published in Microbiology
As humans, we strongly depend on our bacterial flora and their products for our continous good health. On the other hand, bacterial infections remain an important cause of disease and mortality worldwide, and the spreading of antibiotic resistance has the potential to further complicate and worsen infections considerably. Novel antibiotics must and will be developed, but resistances are bound to appear against these novel drugs as well. A supplementary approach lies in the targeted activation of the host’s immune system – be it through vaccination, monoclonal antibody or other recombinant interventions. Indeed, the use of the efficient host’s immune system to overcome bacterial infections might add benefit to any anti-bacterial therapy, and we aimed to develop new methodology to study anti-bacterial antibody repertoires with single-antibody and biophyisical resolution within this work.
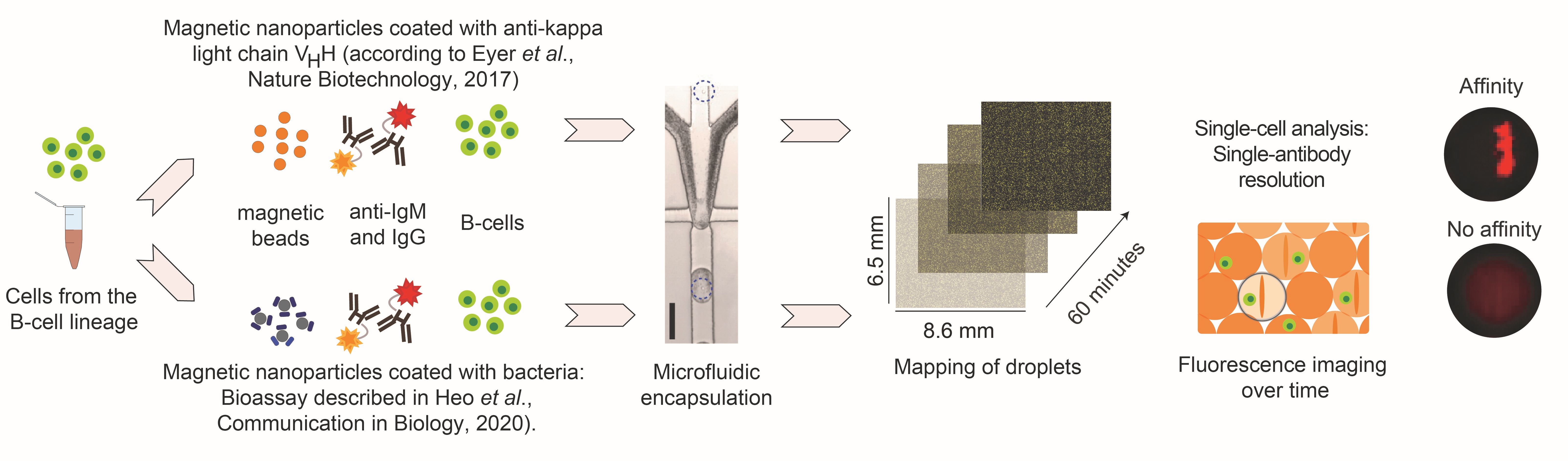 Fig. 1: Overview of the employed droplet experiments used to study anti-bacterial immunizations. Here, we extracted cells from the B-cell lineage from immunized mice, and encapsulated these cells either with
Fig. 1: Overview of the employed droplet experiments used to study anti-bacterial immunizations. Here, we extracted cells from the B-cell lineage from immunized mice, and encapsulated these cells either with
i) a bioassay that allows to measure antibody secretion and the total frequency of IgG- and IgM-SCs (beadline assay, regardless of recognized antigen, top), or
ii) a bioassay to specifically measure anti-bacterial antibodies (bactoline assay, bottom).
Individual cells are encapsulated with the respective assays, the droplets mapped in 2-dimensions using our custom microfluidic devices and antibody secretion and binding is measured over time. For more information, please refer to Heo et al., Communications Biology, 2020, or Bounab, Eyer et al., Nature Protocols, 2020.
First, we had to develop a bioassay that achieves single-antibody resolution, but also allows us to extract additional, appropriate biophysical information about each individual interaction. Here, we identified the need to quantify the number of available and accessible bacterial epitopes (an important parameter for eventual secondary antibody effects) and the affinity of the antibody against the target epitope. Therefore, we started with the development of such an assay, and after characterization and validation of the assay we moved towards our first immunization experiments.
After these initial experiments, we had several discussions and suddenly realized that we had neglected the most important parameter in our considerations so far - namely specificity. We did not quantify whether our induced antibody repertoires were specific for the bacteria used for immunization; or whether the induced antibodies would also bind closely related, related or unrelated bacteria. Indeed, induced anti-bacterial antibodies need to be highly specific towards the infectious agent to be clinically useful (otherwise they would simply bind to almost all present bacteria, and disturb the normal flora). We were wondering whether bacterial specificity is actually the standard case, or whether antibacterial antibodies are highly cross-reactive. Arguments for both cases can be made - certain bacterial epitopes are very similar in their structure, and cross-reactivity of antibodies has to be expected. On the other side, there are also specific bacterial epitopes only present in distinct species and even strains - making antibodies that target these epitopes highly specific. While our answer is far from conclusive, we found substantial differences within the affinity/epitope profiles in mice immunized with various species of heat-killed bacteria; and in their potential to cross-react with various other bacteria. Indeed, some of our immunizations revealed high cross-reactivity of the secreted IgG repertoires, binding to even unrelated bacteria with high affinity. Interestingly, such cross-reactivity was also observed in various commercial antibodies. The full story can be found here.
In light of these findings, the question remains whether immunization or antibody-based therapy represents a viable supplementary option to fight antibiotic resistant infections. In this regard, we are currently trying to understand the reason behind this cross-reactivity from an immunological perspective using different models and bacteria, and to develop strategies to achieve a more focused response.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in