One Health meets network science: A quantitative approach to zoonotic diseases
Published in Ecology & Evolution, Microbiology, and Public Health

The challenge of characterising zoonotic interfaces
One of the core difficulties in managing zoonotic diseases, those that jump between animals and humans, is accurately characterising the interfaces where cross-species transmission occurs. The project began with my question to Anna E. Vogl for her Master's thesis at the University of Veterinary Medicine Vienna, Austria: "Could we characterise zoonotic interfaces in Austria and visualise them?" We aimed to explore the human-animal-environment transmission contexts where zoonotic agents could spillover or be exchanged between hosts and the environment. We found that studies rarely specified which interfaces they examined. As parasitologist Anja Joachim pointed out, simply studying the presence of a parasite in cat faeces (like Toxoplasma) doesn't tell the whole story. Are we looking at the cat-environment, environment-human, or cat-human interface?". The concept of “interface” remained unclear. This motivated us to develop a novel approach to zoonoses and demonstrate it through a case study.
The data
We systematically compiled a dataset of documented zoonotic interactions spanning 47 years of scientific publications and national laboratory reports from Austria. By “interactions”, we mean when zoonotic agents are evidenced in animal hosts, vectors, environments, or foodstuffs. We met here an important challenge: data quality. We realised that species were often inaccurately named or had undergone name changes over time. Authors sometimes referred to sampled animals generically as “duck” or “lizard”, without specifying the species. Food samples were similarly vague, often described simply as “food of animal origin” without specifying details. Cleaning the data and validating the common and scientific names of animals, invertebrates, and zoonotic agents, required weeks of effort to ensure accuracy. Additionally, as co-author Annemarie Käsbohrer remarked, zoonotic agent names posed challenges. For example, shiga toxin-producing Escherichia coli (STEC) strains could refer to both verotoxigenic E. coli (VTEC) and enterohaemorrhagic E. coli (EHEC).
Ultimately, the final dataset includes both zoonotic interactions and epidemiological information on the reported cases. Notably, it revealed the emergence of (at least) eight zoonotic agents in Austria between 1975 and 2022, including West Nile and Usutu viruses, indicating an average of one new zoonotic disease every six years.
Integrating One Health and network science to study zoonotic interactions
Approaches to host-pathogen interactions have used network science, in which the networks share a common data structure, using a connection (called “edge”) list that reflects host-pathogen relationships1. However, there is a notable gap in these approaches when it comes to zoonotic systems, as most zoonotic agents persist in multiple compartments. Besides vertebrate hosts (among which, humans), these compartments include food, invertebrate vectors (like mosquitoes or ticks), and the environment (built or natural, like contaminated water), which are typically absent from conventional host-pathogen networks.
As a veterinary epidemiologist familiar with the One Health concept2, I recognise that quantifying risk at the human-animal-environment nexus is a major challenge for the One Health community. Having worked at the Complexity Science Hub for almost five years, inspired by the quantitative approaches to complex problems, I saw the potential to bridge the gap. This led us to utilise the full dataset and adopt a systems-thinking approach. We created a real-world network that mapped the connections between two different types of actors (think “points” in the network): (i) the zoonotic sources, including vertebrate hosts, arthropod vectors, environmental matrices, and foodstuffs, and (ii) the zoonotic agents, i.e., viruses, bacteria, and eukaryotes. Think of it like a food web, but instead of focusing on energy flow, this "zoonotic web" represents transmission of infectious agents.
By incorporating different types of zoonotic sources and agents in a single network, we introduced complexity as well as challenges in analysing the data and interpreting the results from an epidemiological standpoint. However, the approach provides a more holistic picture of potential zoonotic transmission chains that aligns with the One Health framework.
We addressed the limitations of bipartite networks (“bipartite” reflects that it represents connections between two distinct sets of points) by projecting it into a simpler unipartite network, focusing on connections between zoonotic sources, weighted based on the number of zoonotic agents they shared. To account for research biases, we adjusted these weights to reflect research effort using the number of investigations per zoonotic source. The network could then be explored using traditional methods from social network analysis.
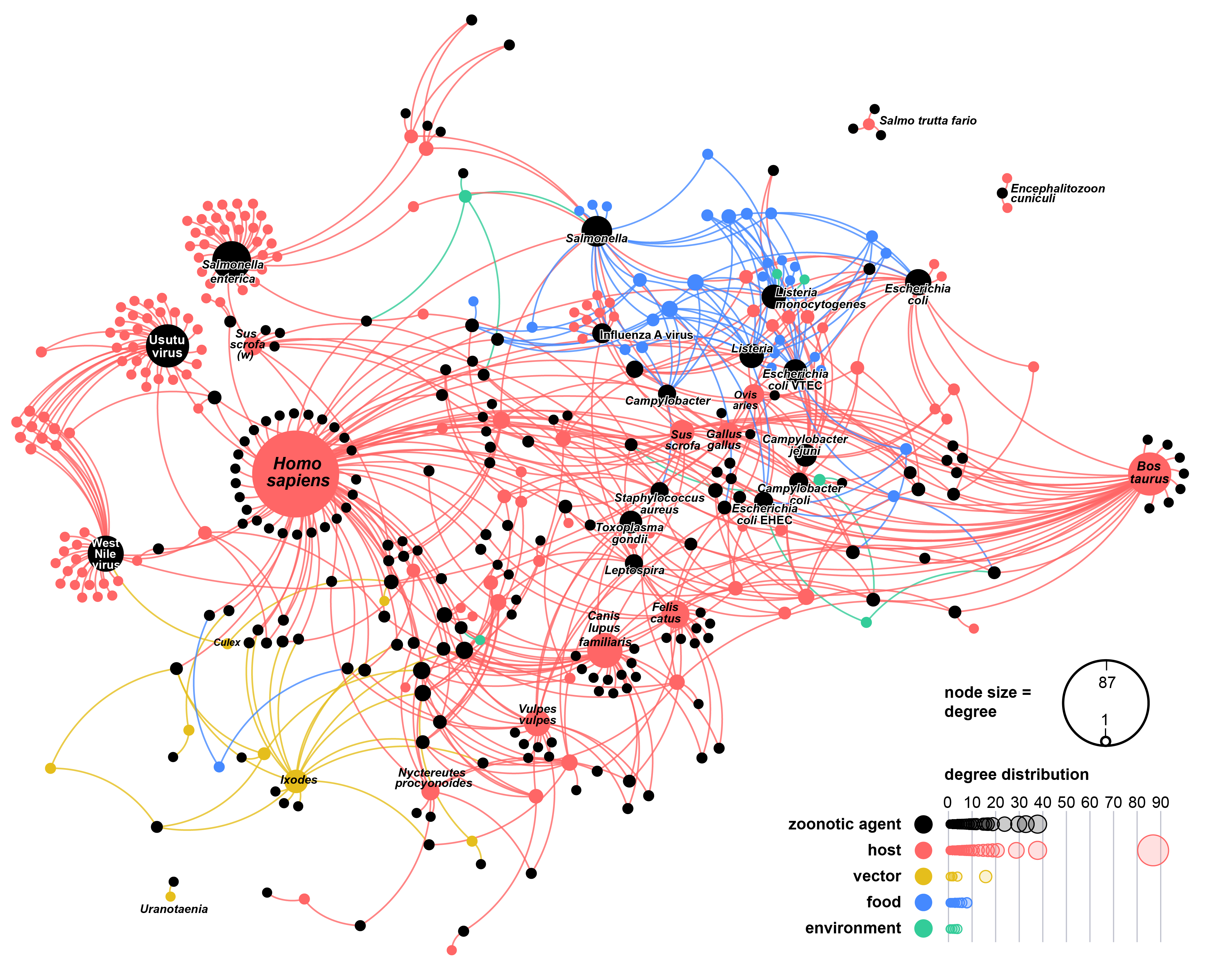
In Austria, our network analysis revealed that certain animals and food sources are more likely to be involved in the spread of zoonotic diseases. For example, chickens, cattle, and meat products appeared to be particularly important. We identified six communities, or “clusters”, of zoonotic agent sharing. Of these, the community involving humans, domesticated (e.g., dogs, cats, cattle, pigs), game (e.g., foxes, cervids), and synanthropic species (e.g., Norway rats, house mice) exhibited the highest overlap of zoonotic agents. This indicates that this community poses the highest risk for zoonotic spillover.
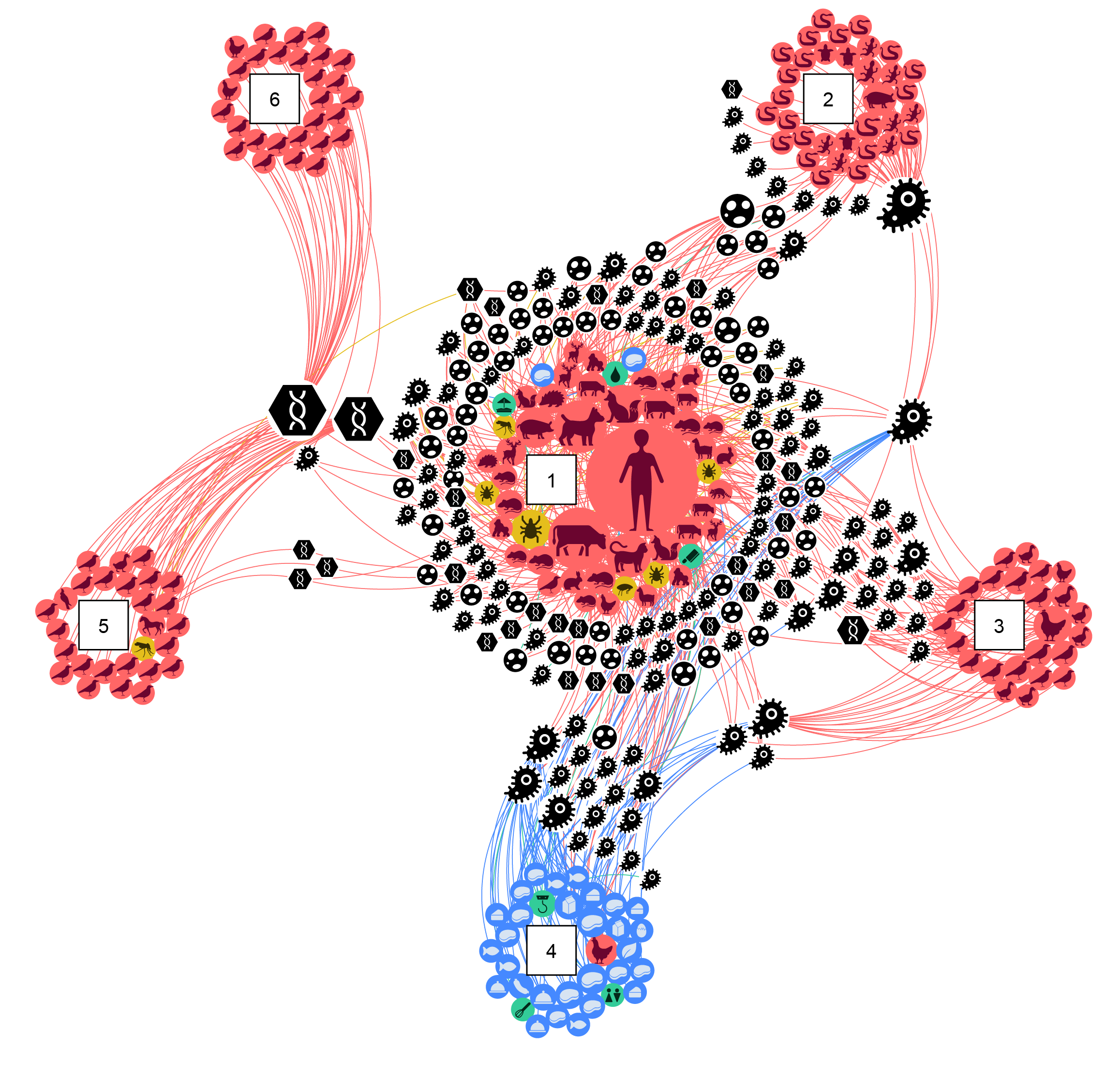
(numbers in squares represent communities of zoonotic agent sharing)
The "zoonotic web" was a great start, but there was still a piece missing. I wanted to make sure this approach truly captured One Health interfaces. Seeking further insights, we teamed up with Gavrila Amadea Puspitarani, who introduced a fresh perspective centred on identifying specific network structures known as 3-cliques, i.e., triangles formed by a set of three “points” all connected to each other. She specifically searched for triangles involving each compartment of the traditional One Health triad (humans, animals, and the environment). We named these structures "One Health 3-cliques”; they enable quantifying and ranking One Health interfaces, i.e., identifying “hotspots” for zoonotic agent circulation. In Austria, we found an increased co-occurrence of zoonotic agents at human-cattle and human-food interfaces, suggesting an elevated likelihood of zoonotic spillover.
Visualising our results to raise awareness
Sharing our research was not just about publishing a paper. We wanted to make this knowledge accessible to everyone, spark interest, and raise public awareness about zoonotic diseases. Liuhuaying Yang, our visualisation expert, transformed complex network data into clear, interactive visuals. To make it more accessible, she has added a search function, following the advice of our communication team, Anja Böck and Eliza Muto. This allows users to explore zoonotic sources and agents, seeing how they connect in the network. By making this information accessible, we aim to encourage behavioural changes that can help prevent zoonotic spillover at source.
Interdisciplinarity and communication
Throughout this project, we engaged in extensive discussions to make sense of the data and integrate perspectives from various disciplines. Our approach, rooted in the One Health concept and network science, emerged from interdisciplinary dialogues between veterinarians, data scientists, and visualisation experts. By overcoming language barriers, including those in programming, these fruitful discussions enabled us to translate quantitative findings into actionable insights. We envision the replicability of this study in other regions and countries. The more data we incorporate, and the higher its quality, the richer the picture becomes.
References
- Runghen R., Poulin R., Monlleó-Borrull C., Llopis-Belenguer C. (2021) Network analysis: ten years shining light on host–parasite interactions. Trends Parasitol, 37(5): 445-455. doi: https://doi.org/10.1016/j.pt.2021.01.005.
- Adisasmito W.B., Almuhairi S., Behravesh C.B., et al. (2022) One Health: a new definition for a sustainable and healthy future. PLoS Pathog, 18(6). doi: https://doi.org/10.1371/journal.ppat.1010537.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in