Opposite genetic adaptation to the diets of European hunter-gatherers and farmers
Published in Ecology & Evolution

Identifying genetic adaptation to historical diets is a recent research focus in the Keinan lab at Cornell University. Back in April 2016, one of our publications swept the media under the name of “vegetarian gene”. In that study, we found that an insertion-deletion (indel) polymorphism in a fatty acid desaturase gene, FADS2, was under positive selection in South Asia, Africa and parts of East Asia. The insertion allele was adaptive to the local plant-based diets by enhancing the biosynthesis of long-chain polyunsaturated fatty acids (LCPUFAs) from plant-derived short-chain precursors. We did not find selection signals in Europeans. This puzzled us because Europeans, like other farming populations, relied heavily on plant-based diets. One possible explanation is the long history of milk drinking in Europe. But we suspected that our original tests might not have enough power. We set out to examine the presence of positive selection on this indel in Europeans with new datasets and methods.
We started by applying a new ancient DNA (aDNA)-based selection test developed by Iain Mathieson with David Reich and colleagues, who also for the first time reported the presence of positive selection on FADS genes in Europe (Iain wrote a follow-up blog post speculating possible evolutionary scenarios). Our original idea was simple: we would impute (computationally infer) the indel into aDNA and then apply the same aDNA-based test. After a deep dive into the aDNA literature and a few days of data crunching, we got the result: no signal for the indel. So, our original idea failed (In hindsight, we didn't have high qualify reference data for imputing this indel. More research to come addressing this issue). However, something unexpected appeared: while many variants in a long genomic region carry significant selection signals, an imputed variant has the strongest signals. It seemed that with imputation we “fine-mapped” the selection signal. We went back to check results from classic selection tests based solely on modern samples and were very excited to find the same refined peak. Meanwhile, we noticed that there is a South-North frequency gradient for the peak variant and for other variants in the genomic region: higher adaptive allele frequency towards southern Europe (Fig. 1). Could there be geographically different selection pressure? Indeed, our formal evaluations suggest that either selection was stronger or it started earlier in southern Europe.
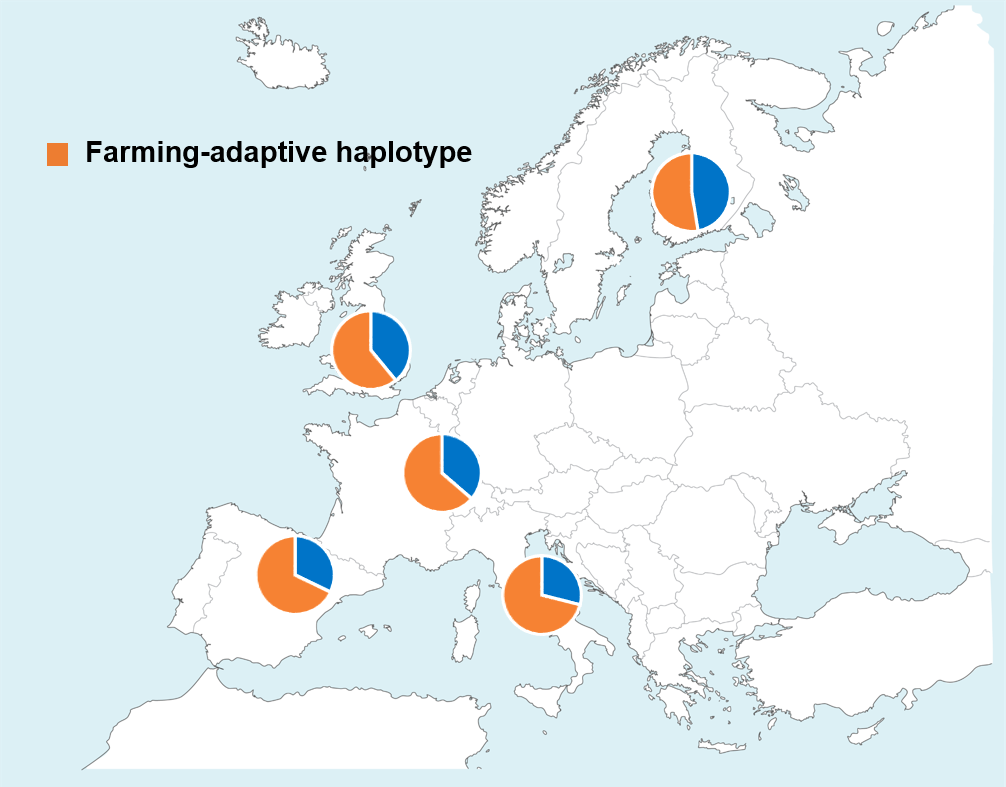
Fig. 1. Geographical frequency distribution of the farming-adaptive haplotype in Europe. The haplotype is formed by multiple FADS variants that are in linkage disequilibrium over a long genomic region.
While we had been working on ancient samples dated from 8,500 to 2,500 years ago (after the Neolithic/Agricultural revolution), a new set of much older ancient samples became available in the summer of 2016. They were Ice Age European hunter-gatherers dated from ~45,000 to 7,000 years ago. Examining FADS genes in these samples, we found something dramatic: the frequency of the farming-adaptive haplotype decreased over time in hunter-gatherers, while the frequency of another haplotype of the same variants (hunting-adaptive haplotype) increased over time in hunter-gatherers and then decreased after the Neolithic revolution in European farmers (Fig. 2). With the help of Montgomery Slatkin and Joshua Schraiber in running their software to infer selection based on allele frequency time series, we confirmed the presence of positive selection on the same FADS variants in hunter-gatherers, but on alleles opposite to those adaptive in farmers. The selection pattern on some variants in pre-Neolithic European hunter-gatherers is similar to what was observed before in Greenlandic Inuit, who have a seafood-based diet. Further association analysis with gene expression and lipid levels revealed that farming-adaptive alleles increase FADS1 expression and enhance LCPUFAs biosynthesis, while hunting-adaptive alleles have opposite effects.
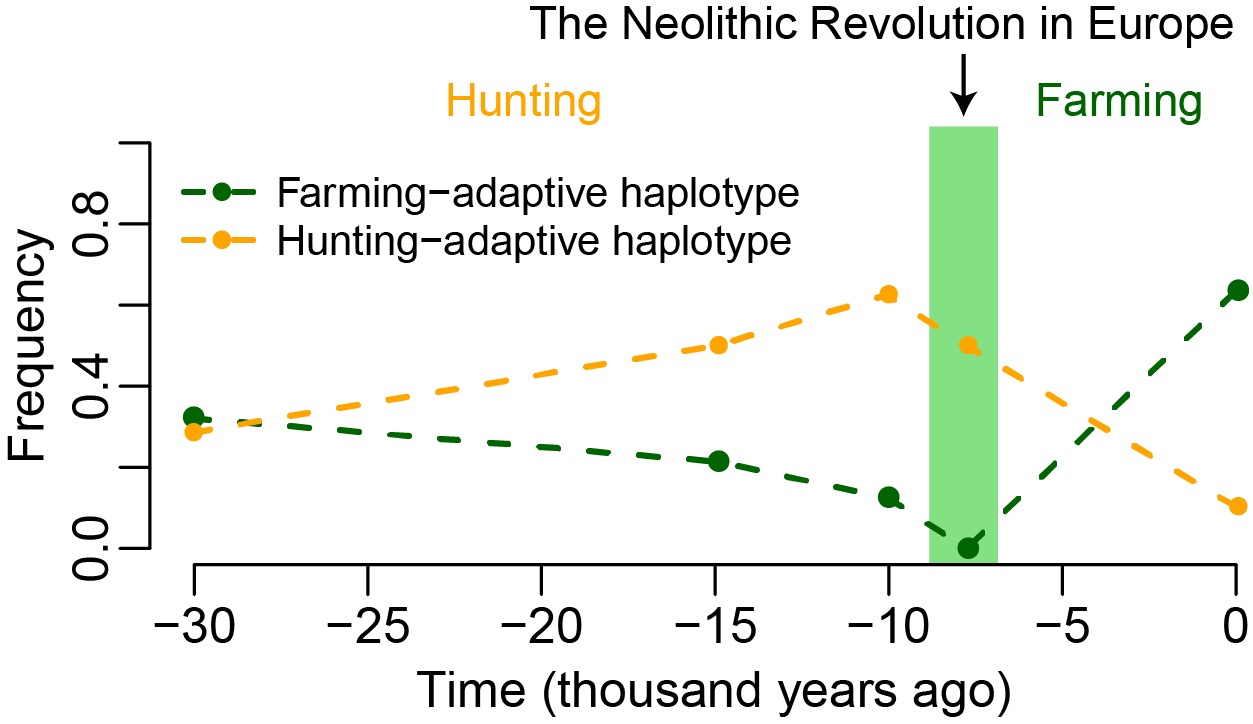
Fig.2. Hunting-adaptive haplotype increased in frequency before the Neolithic revolution but decreased in frequency afterwards, while the trend for farming-adaptive haplotype is opposite. The two haplotypes were defined with opposite alleles of the same variants in FADS genes.
As these evolutionary patterns and functional mechanism became clear, we could not help but ask: what was the underlying selective force? We were fairly sure about the role of plant-based diets for recent farmers. But was there stronger reliance on domesticated plants in southern European farmers? Regarding pre-Neolithic hunter-gatherers, did they have a diet similar to that of Greenlandic Inuit? Fortunately, Ofer Bar-Yosef, an anthropologist at Harvard University, was willing to join our project and referred to anthropological findings that support our evolutionary observations. Indeed, pre-Neolithic hunter-gatherers had an animal-based diet with substantial aquatic contribution. After the Neolithic revolution, all farmers relied heavily on plant-based diets, but the reliance was stronger in the South because northern farmers had more access to aquatic food from the Atlantic Ocean and the Baltic sea and they also drank more milk.
Putting all these evidence together, we finished the first version of our manuscript on a very special night – Nov 8, 2016. We submitted it the next day. On the same day, a manuscript from the research group of Rasmus Nielsen was posted on bioRxiv. While tempting, we made a conscious effort not to read this manuscript until ours was accepted in order to avoid bias. When we finally got to read their manuscript, we were very pleased to find that it independently confirmed some of our observations. It also found that some of the variants we studies have been under recent positive selection after the Neolithic revolution (or since the Bronze Age). Our paper further revealed the unique pattern in pre-Neolithic hunter-gatherers.
Our paper in Nature Ecology & Evolution could be found here. A read-only version of the full-text could be found here.
This post was written jointly by Kaixiong Ye and Alon Keinan.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in