Can we model the benchmark of human embryonic development on a petri dish? This has been the fundamental question in developmental biology. Partition of blood, brain, heart, kidney, gut, and placenta is the major benchmark of human embryos. This study achieved the partition of blood, heart, and placenta in human embryonic organoids.
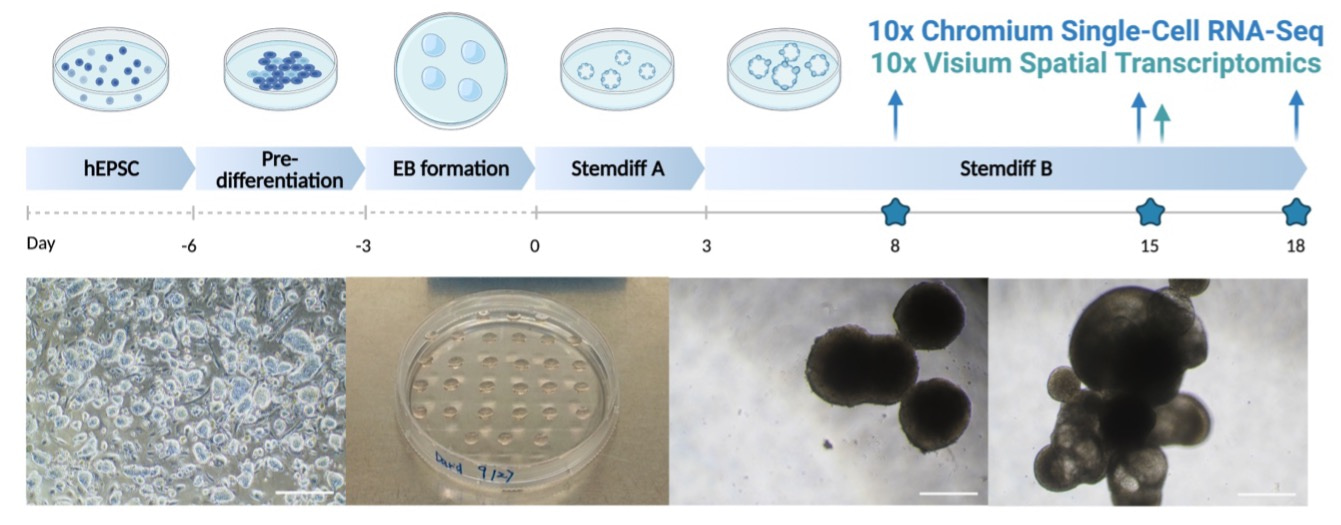
We believe in the power of stem cells. They could build embryo-like structures on a petri dish. This is how our work started. We adapted highly potent stem cells, namely expanded potential stem cells. We leveraged the self-organizing capacity of stem cells and generated three germ layer patterning. We named them Human EMbryonic Organoids (HEMOs).
HEMOs recapitulated fundamental events in human embryos. Through morphogens and cytokines exposure, HEMOs generated cardiac precursors, neural crest, blood, placenta and yolk sac. These correspond to the trunk region of human embryos. Given the technological advance of high throughput sequencing methods, we exploited the time series single-cell (sc)RNA-sequencing and spatial transcriptomics to study cell-cell interactions in the development of embryonic tissues.
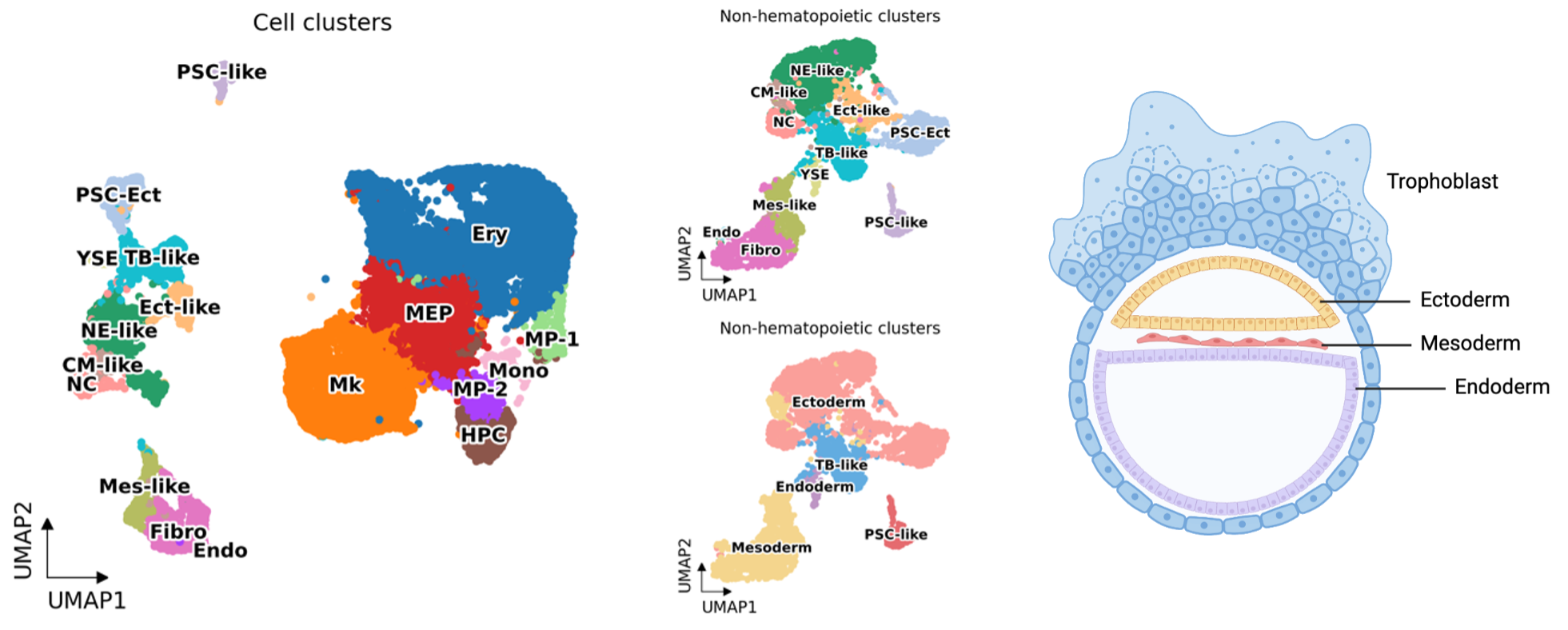
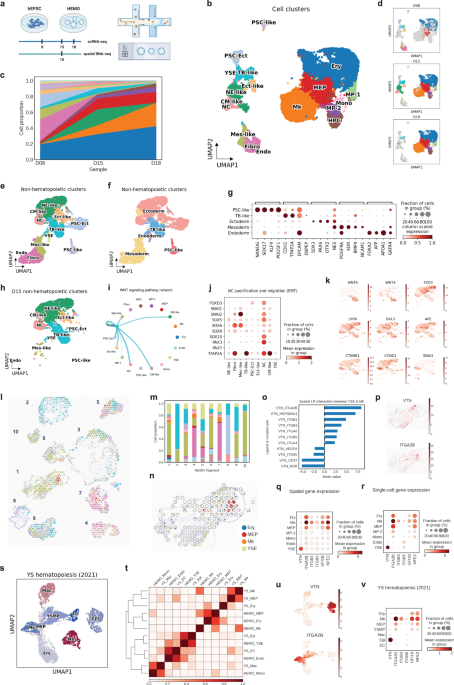
HEMO followed human embryonic development. Among the cells from scRNA-seq, we annotated eighteen cell clusters, including human developmental lineages and hematopoiesis. HEMO acquired extra-embryonic lineages (trophoblast-like tissues, yolk sac endoderm), ectoderm (pluripotent stem cells with ectoderm specification, ectoderm-like cells, neural ectoderm-like cells, neural crest), and mesoderm (mesoderm-like cells, cardiac mesoderm-like cells, fibroblasts, endothelium).
We addressed cell-cell communication during human embryonic development. During the development of embryonic trunk tissues, we found that WNT signaling from trophoblast (TB)-like tissues was predominantly connected to neural crest populations. The high expression of neural crest specification genes (FOXD3, SOX5, SOX6, SOX9, SOX10, PAX3, PAX7, TFAP2A) and migration genes (SNAI1, SNAI2) suggested the maturation and migration of neural crest cells. These observations suggest a potential role of TB-like tissues in promoting neural crest maturation and migration in the HEMO.
We defined spatiotemporal gene expression landscape in human embryonic tissue specification. Now scRNA-seq is mature, still integrating them with spatial information is highly demanded. We collaborated with a mathematician Yang Can at Hong Kong University of Science and Technology. He generated an algorithm that feed both scRNA-seq and 10X Visium dataset, called SpatialScope. We achieved near single-cell resolution information of topology and gene expression in slices of HEMOs.
We defined the embryonic hematopoietic niche in the yolk sac and their potential key signal. In the spatial slices, we consistently observed yolk sac erythro-megakaryopoietic niche, where co-localization of the yolk sac endoderm (YSE), erythroid cells (Ery) and megakaryocytes (Mk) populations frequently happening in the same spots, suggesting physical cell-cell interactions. With the CellPhoneDB, we observed a strong interactive score between YSE and Mk through vitronectin (VTN) signaling, with VTN-ITGA2B ranked highest. VTN was exclusively expressed in YSE, while ITGA2B was expressed in the erythro-megakaryopoietic populations, with the highest expression level in Mk. Other integrin subtypes ITGB3 and ITGB5 as receptors of vitronectin were highly expressed in Mk. Consistently, vitronectin-integrin genes revealed the similar pattern in our paired scRNA-seq dataset and spatial transcriptomics. These findings indicate that yolk sac endodermal cells are the primary source of vitronectin expression, and that the integrin pathway plays a role in promoting megakaryopoiesis in HEMOs.
How close our HEMO with the real-world? To validate our findings, we compared HEMOs with the scRNA-seq dataset of human fetal tissue to examine the physiological relevance (herein called ‘YS hematopoiesis’). In the YS hematopoiesis dataset, VTN was mainly expressed in yolk sac epithelial cells, and ITGA2B was mainly expressed in megakaryocytes. Both YS hematopoiesis and HEMO achieved consistently similar expression patterns of vitronectin, integrins, GFI1B, and NFE2. Taken together, it evidences that HEMO could mimic human embryonic hematopoiesis in the yolk sac, and vitronectin-integrin signaling as a molecular signature of megakaryopoiesis.
In summary, stem-cell-derived embryos advanced understanding of human embryonic development. This offers a platform to perturb and engineer genes important for human development as well as disease modelling. By adopting the defined medium in the commercial kits, we enhanced the reproducibility of the techniques between different labs.
The possible application of HEMO is to engineer hematopoietic and immune cells. The contribution of extraembryonic tissue yolk sac in megakaryopoiesis through vitronectin-integrin axis holds a promise to generate platelets from stem cells for cell therapies. The generation and engineering of monocytes and macrophages could target cancers and infectious diseases. The spatial understanding of the yolk sac hematopoietic niche and leveraging their factors could promote the generation of immune cells from stem cells.
Follow the Topic
-
Signal Transduction and Targeted Therapy

This is an international, peer-reviewed, open-access journal publishing articles related to signal transduction in physiological and pathological processes, alongside signal transduction-targeted therapeutics in the form of biological agents and small molecular drugs used to treat human diseases.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in