OXPHOS remodeling in high-grade prostate cancer involves mtDNA mutations and increased succinate oxidation
Published in Cancer
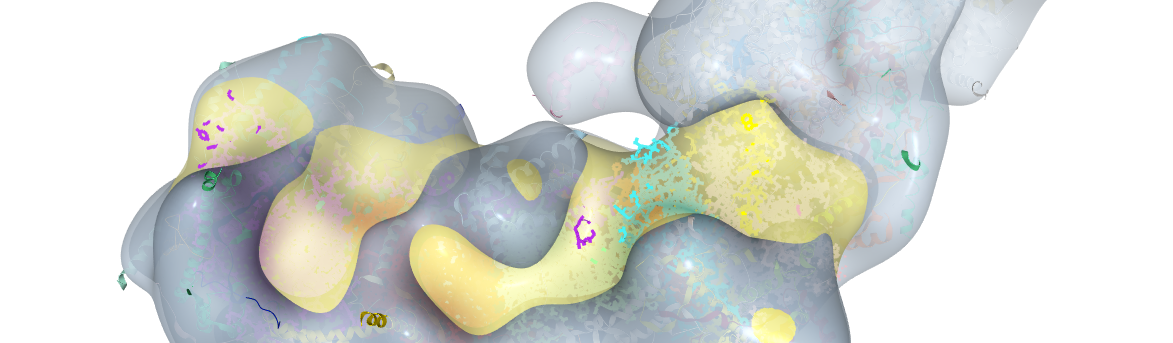
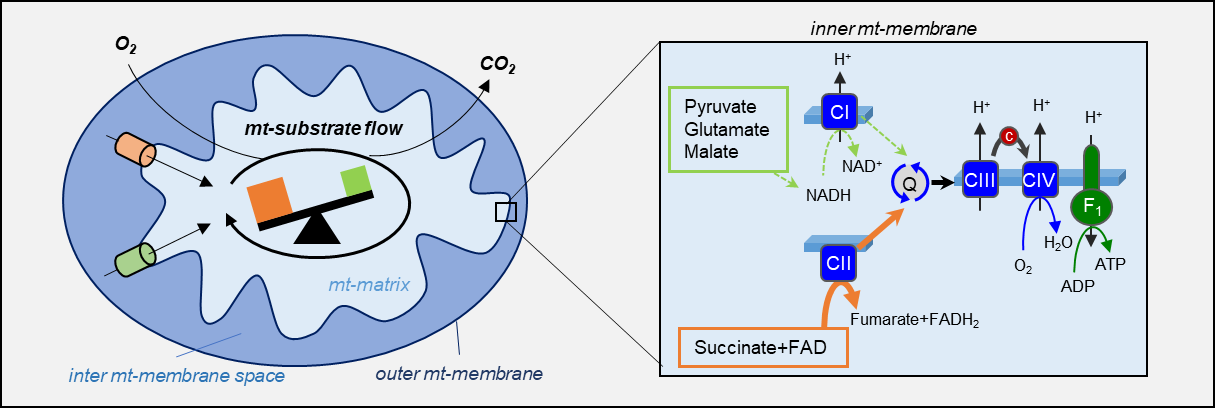
Fig. 1. OXPHOS substrate shift from glutamate and malate to succinate in high-grade prostate cancer tissue
Cellular metabolism is a well-balanced act. Each cell must ensure that enough metabolites are burned for energy production, while simultaneously sufficient metabolites are spared to serve as bio-precursors for cellular repair and signaling purposes. In cancer cells, this balancing-act is suspected to be skewed in a specific direction to sustain uncontrolled growth. In these cases, cancer cells might take advantage of non-canonical substrate pathways to satisfy specific substrate needs. This can be accomplished by a phenomenon of metabolic reprogramming that is best described as a process of “fuel partitioning”. Considering that the mitochondria are the powerhouses of the cell, we hypothesized that fuel partitioning is likely to affect cancer cell specific mitochondrial substrate pathways in a detectable way.
Mitochondria produce the “energy current” ATP by a stepwise oxidation of various substrates during a process called oxidative phosphorylation (OXPHOS) which takes place at the inner mitochondrial membrane. During OXPHOS, electrons resulting from metabolite oxidation are fueled into a mega-complex called “electron-transfer” (ET) system which is composed of a battery of serially connected protein complexes. Electrons enter the ET-system either via Complex I (CI) or Complex II (CII), depending on substrate type. Because the process of OXPHOS consumes oxygen, we can monitor the mitochondrial turnover of cellular substrates in a sample by measurement of oxygen changes in a closed system via a method called High-Resolution Respirometry (HRR). We recently developed a method to assess OXPHOS via HRR in native prostate tissue samples to measure the effect of various substrates on mitochondrial energy production using an instrument that was developed, and is still produced, here in Innsbruck.
In the present study, we applied our method to measure OXPHOS in paired benign and malignant prostate tissue samples to evaluate the presence and extend of fuel partitioning in primary prostate cancer (PCa). Interestingly, we found that PCa tissue shows a high preference for succinate (which is oxidized via CII) to compensate for a decrease in the capacity to oxidize substrates via CI. To our surprise, this effect was mainly present in high-grade tumors, indicating that initiation of fuel partitioning might be a prognostic indicator. Consequently, the next step for us was to identify potential causes or mechanisms linked to this metabolic reprogramming.
We first looked at the mitochondrial DNA (mtDNA), a small genome located within the mitochondrial matrix, since some ET-proteins are encoded by mtDNA. It is known that non-synonymous mutations in mtDNA genes coding for CI subunits can cause a decrease in the capacity to oxidize substrates linked to CI. It has also been reported that cancer tissue exhibits high numbers of mtDNA mutations. We therefore thought that, at least in some cases, the decrease in CI activity might be caused by non-synonymous mutations that would disrupt CI protein stability and/or activity. We performed high-throughput Next-Generation Sequencing (NGS) on the Ion Torrent Proton (>10,000x) in 3 different runs. We applied our mtDNA NGS data analysis software (mtDNA-Server) to screen for point mutations. Sequencing of our samples indeed uncovered high loads of potentially deleterious mtDNA mutations. We also found that samples harboring such mutations with high allele frequencies showed a marked decrease in CI activity in HRR. To substantiate our functional findings, we evaluated the impact of these mutations in-silico using a 3D model of CI. To make our findings accessible to all scientists, we posted the 3D model including the locations and types of the mutations on GitHub and our website. Additionally, we analyzed the mitochondrial copy number via qPCR and estimated the mitochondrial mass by immunohistochemistry. Both parameters were found increased in the high-grade tumors.
In a second step, we looked at mRNA expression patterns of key enzymes, which are involved in the metabolic pathways providing substrates for OXPHOS. Based on our HRR findings, we hypothesized that expression patterns of proteins and enzymes that regulate fuel partitioning might be altered in tumor samples. Not surprisingly, transcriptome NGS of paired samples revealed that mRNA expression of enzymes and transporter proteins involved in succinate metabolism were upregulated. Since our colleagues from the University of Salzburg recently established immunohistochemical assays to asses ET protein expression, we used their method to study Complexes I and II. We found elevated CII marker protein levels associated with tumors harboring deleterious CI gene mutations
In a final step, we wondered if the mRNA expression patterns observed in the samples characterized by massive fuel partitioning might have prognostic potential. For this, we extracted a selected number of genes that were highly representative for this metabolic phenotype and used publicly available PCa cohorts to look for differences in disease outcome in patients that were characterized by this gene signature. Interestingly, we found a consistent trend towards worse outcome in those patients whose PCa was characterized by this gene signature. This might indicate a prognostic significance of the fuel partitioning we detected in PCa.
While our results provide some initial insights on the process of fuel partitioning in PCa and some potential causes for this re-programming, we believe that these pivotal findings are only the beginning. Our study raises important questions that we must answer to get a full understanding of the metabolic adaptation in human cancers. We hope that, in the end, this understanding might help to develop novel therapeutic strategies that will improve the lives of cancer patients in the future.
Written by Bernd Schöpf, with adaptations from Hansi Weissensteiner and Helmut Klocker
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in