Pancreas and Pancreatic Cancer ExoConnectomes
Published in Cancer, Protocols & Methods, and Cell & Molecular Biology
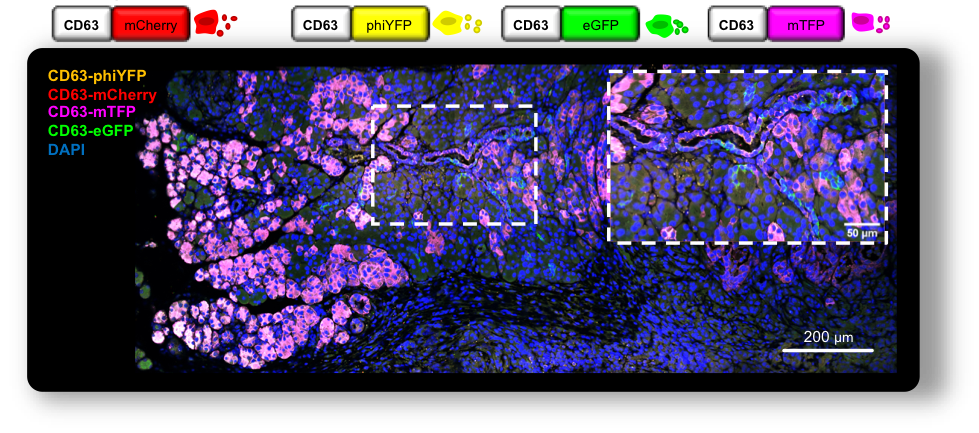
Recent advances in the development of tailored mouse models for in vivo tracking of exosomes have opened new avenues to uncover their significance in different biological scenarios. Models that closely mimic the biological system can help consolidate our existing knowledge of the functions of exosomes but also uncover features of these vesicles. This improves our understanding of the overlooked homeostatic processes but also sheds light on the pathophysiology of cancer. Consequently, these findings hold potential for improving patient care by providing insights into the biology of the disease and by advancing the state of the art in exosomes research, enabling their use as potent therapeutic vehicles or targets.
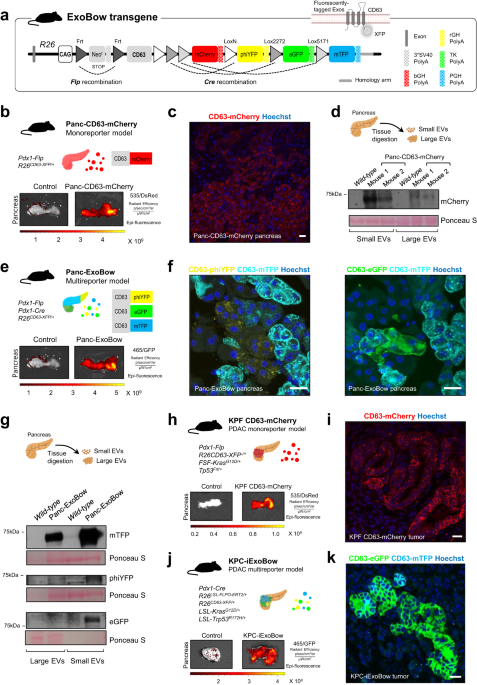
In this study, we developed a versatile genetic approach using a CD63-based reporter mouse model to track exosomes specifically from pancreas cells. By investigating the distribution of pancreas exosomes during PDAC progression, we made several discoveries that enlighten the biological significance of exosomes in the healthy pancreas and in PDAC. One of our key findings is that the frequency of a particular cell type does not determine the communication routes that take place. This observation suggests that intercellular communication in vivo is not random but rather occurs in a coordinated manner. These findings provide insights into the dynamic interplay between different cell types during cancer progression, uncovering a local and at-a-distance coordinated network of communication that provides a deeper understanding of how PDAC evolves and progresses.
Our work provides evidence for the role of exosomes secreted by the healthy pancreas in restraining angiogenesis. Hence, exosomes could be a means to maintain overall homeostasis and organ fitness. Interestingly, our data show that tumor angiogenesis is also modulated by PDAC exosomes, shedding light on important mechanisms driven by exosomes in PDAC to support disease progression. We also demonstrate communication with CAFs, and CAFs with low expression of αSMA (usually associated with an inflammatory phenotype) are enriched nearby PDAC lesions with proficient exosomes secretion, showcasing the potential role of cancer exosomes in modulating the landscape of CAFs and influencing the prognosis of PDAC tumors. Finally, communication was also detected with immune cells within the healthy pancreas and PDAC, albeit at very low levels.
Inter-organ communication was enhanced in PDAC in comparison to the healthy pancreas. The kidneys and the thymus are the organs where most cancer exosomes accumulate. The molecular mechanisms underlying the observed organotropism and its biological impact on disease progression remain to be elucidated. Regarding communication with organs that host PDAC metastasis, cancer exosomes accumulated in the lungs (both in early and late PDAC stages), yet the same observation was not true for the liver, which is the most frequent site of metastasis.
The integrated analysis of the cargo of PDAC exosomes, combining both protein and RNA content, along with the changes in RNA expression in fibroblasts and endothelial cells following exposure to cancer exosomes, provides valuable insights into the mechanistic underpinnings of observed in vivo phenotypes. This approach underscores the direct and indirect influence of exosomes on target cells, ultimately resulting in their reprogramming and the remodeling of the tumor microenvironment. Furthermore, the concurrent increase in the abundance of exosomes within the PDAC context, compared to a healthy environment, highlights the potential combined impact of both exosomes quantity and cargo diversity in shaping the intricacies of the tumor microenvironment.
Altogether, we have mapped the intra-pancreas and inter-organ distribution of exosomes in the healthy pancreas and PDAC. We demonstrate that communication is not a random event but rather depends on the biological context rather than the prevalence of distinct populations of cells, and that these specific routes of communication that take place in vivo influence the composition of the pancreas microenvironment with a specific impact on angiogenesis and spatial distribution of CAFs. Additionally, we have developed a GEMM to tag and trace exosomes in a lineage-specific manner that can be used in a multitude of biological settings. The broad use of this model in other contexts, pathological and non-pathological, will elucidate on whether the communication routes governed by CD63+ exosomes are specific to the pancreatic context or if more transversal mechanisms are at play. The potential use of this model in its multicolor form further strengthens its versatility and relevance. Dissecting communication routes between subpopulations of the same cell of origin could be crucial to further understanding processes such as homeostasis and intratumor heterogeneity. We believe this study contributes to a better understanding of the biological significance of pancreas exosomes in health and disease.
Although a great effort has been made to improve the current models and methodologies to study exosomes in vivo, no model is bulletproof, and several considerations are in place. First, the nanosize of exosomes represents a major hurdle in their analysis and is limited to the detection threshold of the techniques used. In addition, cells are described to produce heterogenous populations of exosomes, and a consensus marker that includes them all, if existing, is yet to be determined. Specifically, in our work, we study the CD63+ population of exosomes. Even though CD63 is widely described as an exosomes marker, it does not cover all endosomal-derived exosomes, and, on the other hand, it can also be found in other EVs subpopulations. Indeed, the most suitable tetraspanin of choice may vary between tissue/cell-type or even according to the pathophysiological context. Hence, only a comprehensive and coordinated analysis using different models in the same biological setting will provide valuable information to the remaining open questions.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in