Personalized OncoGenomics: A collaboration of scientific and clinical minds at BC Cancer is answering complex questions about cancer and its treatment
Published in Cancer
The seeds were planted for a whole new way to treat cancer, informed by genomics, over a decade ago. Dr. Janessa Laskin, a medical oncologist at BC Cancer asked Dr. Marco Marra, Director of Canada’s Michael Smith Genome Sciences Centre at BC Cancer, if there was anything genomics could do to guide the treatment of one of her patients, who had a rare type of cancer. Armed with next generation DNA sequencing technology, Drs. Marra and Laskin would lead the first study of its kind, to use whole genome and transcriptome sequencing to inform cancer treatment, the results from which were published in a seminal paper in Genome Biology in 2010 (Jones et al., Genome Biol. 2010).

The leaders of BC Cancer’s Personalized OncoGenomics program; from left to right: Drs. Janessa Laskin, Medical Oncologist, Marco Marra and Steven Jones, Directors of Canada’s Michael Smith Genome Sciences Centre, BC Cancer.
Four years later, the Personalized OncoGenomics (POG) was launched. POG, which studies the use of whole genome and transcriptome sequencing to inform patient treatment in advanced cancers, was built upon a shared vision of genomic information to improve cancer care. Making this vision a reality required bringing genomics, oncology, pathology, radiology, hereditary cancer and many other disciplines together. With over 700 adult and 110 pediatric patients sequenced to date, POG has amassed a complete, accurate and permanent dataset of human tumour genomes and transcriptomes (over 200 trillion bases) that has been the backbone for over 40 studies, bringing together a wide range of experts across multiple disciplines in health care and science.
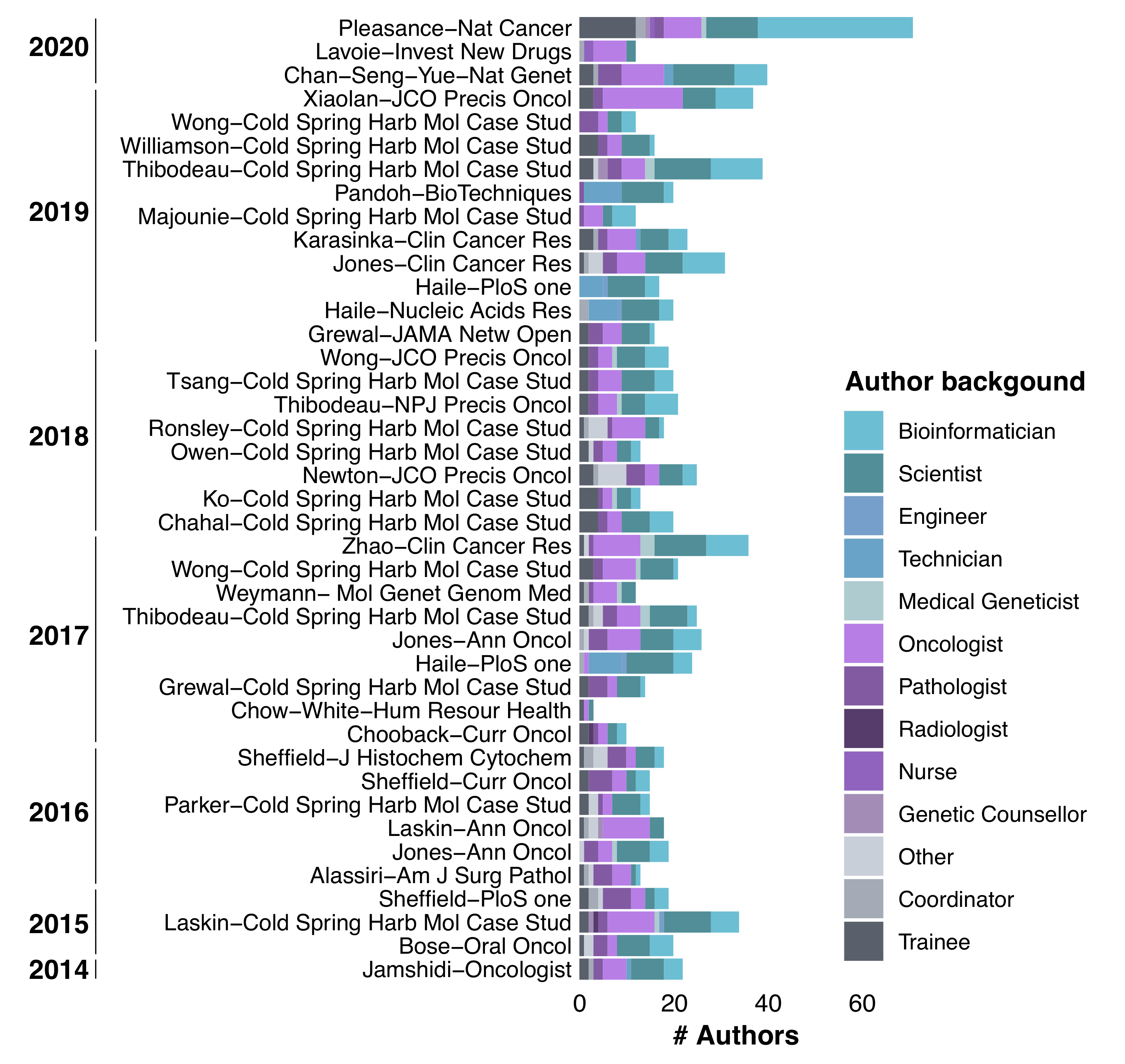
Publications from the Personalized OncoGenomics Program represent contributions from numerous authors spanning a wide variety of backgrounds including bioinformatics, oncology and pathology.
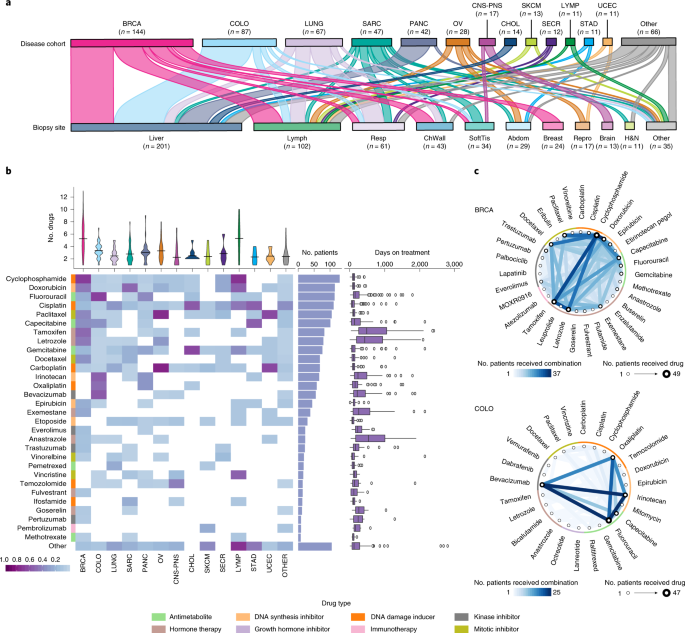
How do advanced cancers differ from primary disease? What are the impacts of prior therapy on the tumour genome, when advanced cancer patients have become treatment resistant? By sequencing advanced, pre-treated tumour samples, and integrating the complete genome and transcriptome sequencing data with patient treatment history and clinical data, POG is generating the data required to begin to answer these questions. Our most recent study, “Pan-cancer analysis of advanced patient tumors reveals interaction between therapy and genomic landscapes” published in Nature Cancer, focused on the impact of therapy on tumour genomes and transcriptomes. This study is our largest collaboration to date and represents seven years of data collection and over two years of data analyses including somatic mutation and copy number calling, structural variant detection, germline analysis, expression clustering, survival analysis and mutation signature discovery. Synthesizing these analyses revealed evidence of acquired treatment resistance due to mutations, copy number alterations and even expression changes, as well as rare non-coding recurrent events that may represent cancer drivers.
The most striking finding was the extent to which treatment exposure was associated with increased somatic mutations in the genome. In fact, patients exposed to platinum-based therapies for more than one year have on average twice as many mutations as those who were not exposed. We also observed specific mutation signatures related to specific drug combinations and tumour DNA repair defects. These findings have both negative and positive implications for patients—the increased mutation burden may provide tumours with more ways to evolve treatment resistance, but may also increase their susceptibility to immunotherapies. This also highlights the need to study advanced and treated cancers, which are currently underrepresented in the field despite having the highest mortality rates.

In long-running, collaborative projects such as POG, recognizing milestones is an important way to celebrate. Today, we’re eager to celebrate the publication of this dataset when we can be together again, as now we’re all working from our respective homes to do our part to reduce the spread of COVID-19.
We are immensely proud of POG, and are very excited to be able to share this new dataset with the research community. As we have demonstrated through our program, collaboration is essential to driving research forward, and broad sharing of both genomic and clinical data is integral to further understanding cancer and improving patient care.
We are very interested to hear how others incorporate our POG data in their research! Please tell us by leaving a comment below.
What started out as a one-off attempt to provide personalized therapy suggestions to a patient in Vancouver has evolved into a deeply collaborative effort that has touched the lives of hundreds of patients and their families, helped launch a new generation of projects bringing genomics to the clinic across Canada, and provided the global community with a rich resource of genomics and clinical data from advanced cancer patients. Together we can work to reduce the burden of cancer worldwide, through research and innovative treatment.
Written by: Laura Williamson, Erin Pleasance and Emma Titmuss
Follow the Topic
-
Nature Cancer

This journal aims to provide a unique forum through which the cancer community will learn about the latest, most significant cancer-related advances across the life, physical, applied and social sciences.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in