Transcriptome analysis reveals key information about a rare pediatric cancer and response to immune checkpoint inhibitor monotherapy
Published in Cancer

Comprehensive genomic and immunological profiling of tumours may reveal unique therapeutic vulnerabilities in rare malignancies. Even when tumour mutation burden is low, studies have shown that immunologically “hot” tumours may respond to immunotherapy. In this study, comprehensive molecular analysis of poorly differentiated chordomas from two pediatric patients revealed key information about this rare cancer with implications for treatment.
The pediatric branch of Personalized OncoGenomics (POG) at BC Cancer Vancouver, Canada—a research study that integrates comprehensive tumour genomic and transcriptomic sequencing into the clinical care of advanced cancer patients—has comprehensively sequenced 148 pediatric tumours and corresponding matched germlines from children with hard to treat cancers. Among these are two pediatric patients with poorly differentiated chordoma, a rare form of sarcoma.
One of these patients was a previously healthy and active eight year-old girl who first noticed some neck pain and stiffness about a year prior to her diagnosis. Her physician requested neck x-rays, which were normal, and she saw a physiotherapist and massage therapist. As the neck pain persisted, she was referred to a pediatrician who requested an MRI of her neck. A mass was found in her C1 vertebral body and a trans-oral needle biopsy was completed. Pathology results—reviewed by multiple experts including Dr. Stephen Yip, a neuro-oncology clinician scientist with BC Cancer—were in keeping with a poorly differentiated chordoma.
As the tumour was unresectable, she started intensive chemotherapy with some disease response. Unfortunately, she had early disease recurrence with a mass affecting the skull base and an enlarging pulmonary nodule, which is when she was enrolled in POG for tumour profiling.
Low mutation burden but high T cell score
Results from whole genome sequencing revealed a tumour almost completely devoid of notable somatic alterations, with the exception of a sole region on chromosome 22 containing a single copy loss of SMARCB1 (INI1). There were no clonal somatic non-synonymous small mutations.
With the absence of genomic variants of interest, we focused our attention on the transcriptome. We observed high expression of the TBXT gene that encodes the transcription factor brachyury—a protein that is characteristically expressed in chordoma—and low expression of SMARCB1.
We also explored the tumour microenvironment using a tool called CIBERSORT1 that deconvolutes bulk RNA sequencing data to estimate the relative abundance of immune cell types in tumour samples. To our surprise, the chordoma sample contained one of the highest CD8+ T cell scores compared to all POG samples that had been sequenced to date.
This led us to look for T cell receptor sequences by interrogating RNA sequencing data with the tool MiXCR.2 This analysis revealed a dominant T cell receptor sequence present in the tumour microenvironment.
We also noted high expression of CD274, the gene that encodes PD-L1, suggesting immune evasion through activation of the immune checkpoint. Based on these results, we postulated that the immunologically hot phenotype might confer response to immune checkpoint inhibition.
Given these results, and a lack of standard therapy options for this patient, her oncologist applied for compassionate access to nivolumab, an immune checkpoint inhibitor with established pediatric dosing. This was well tolerated and follow-up imaging investigations after three cycles of nivolumab monotherapy documented partial response by RANO criteria and decreased mass effect on the adjacent brainstem. Follow up MRIs showed slow progression, however the patient continued on therapy for 14 cycles as she continued to experience clinical benefit with an excellent functional status and quality of life.
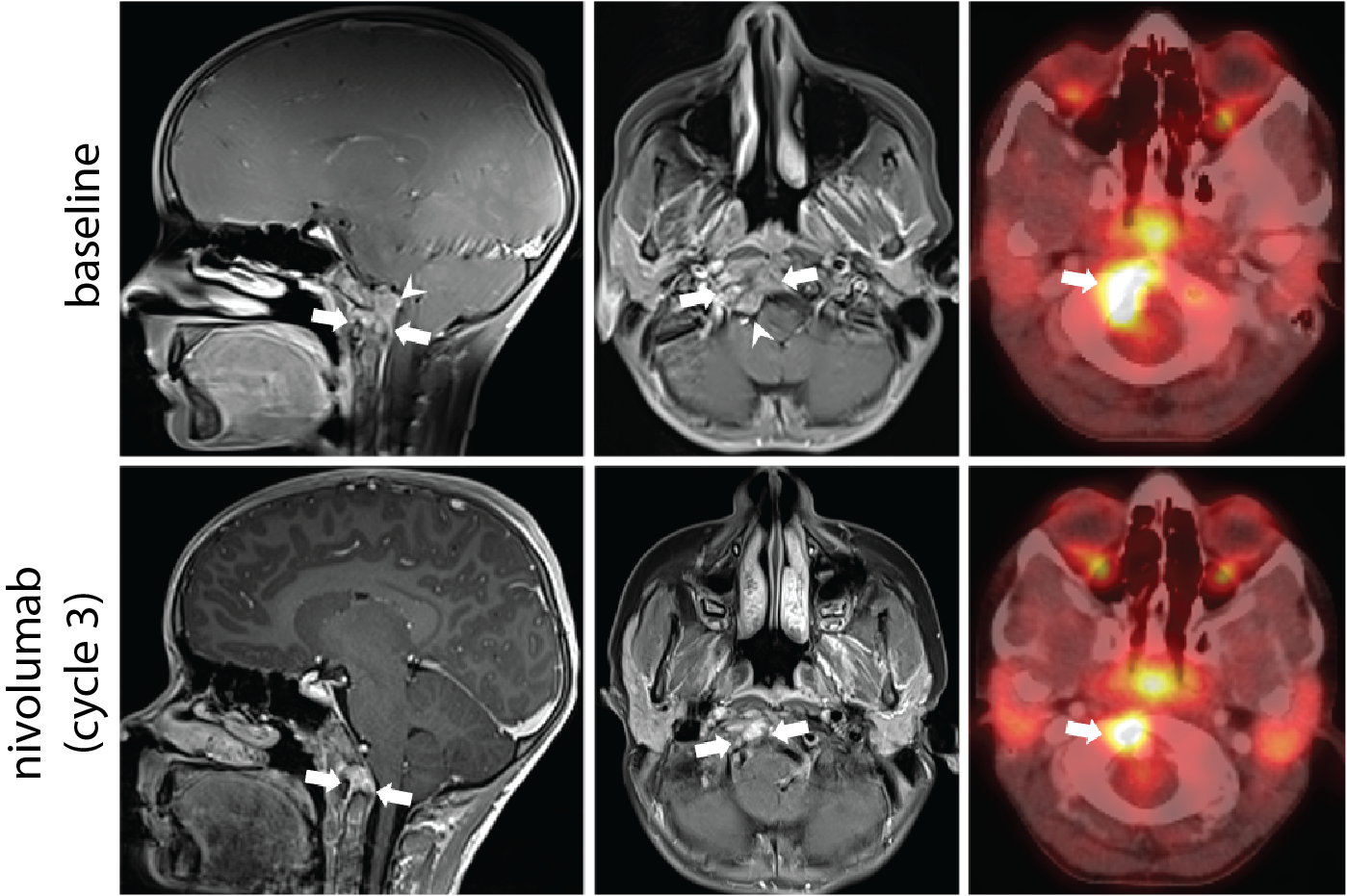
A rare response to immune checkpoint inhibition, and a missed opportunity
This patient achieved disease response to immune checkpoint inhibitor therapy, which is exceedingly rare in unselected cohorts of pediatric solid tumours. This outcome was facilitated by the technology and methodology that was not available to the second patient described in our paper.
The second patient, also diagnosed with poorly differentiated chordoma, was enrolled in pediatric POG several months earlier. Like the first patient described, apart from copy loss affecting SMARBC1, no driver mutations were identified. In both patient samples, CD8+ T cell estimates measured retrospectively revealed a similarly high CD8+ T cell score, indicating that immune checkpoint inhibitor therapy may have also been beneficial in this patient.
Unfortunately, unlike the first patient, immune cell estimates based on deconvolution methods were not discussed at the molecular tumour board as at that time they were not part of our analysis. As a result, immune checkpoint inhibitors were not identified as a potentially beneficial therapy. The patient passed away shortly after sequencing.
It is unclear if immune checkpoint inhibitors would have been a suitable option for this patient and we do not know if they would have had a similar clinical response; however, this missed opportunity highlights the power of incorporating new methodology in precision cancer initiatives, and the value of complete, accurate and permanent datasets amenable to retrospective analysis.
A collaborative approach to understanding mechanisms of drug response
We wanted to understand whether the bioinformatics tools that pointed to a high level of CD8+ T cells, including a clonally dominant T cell clone, were a reliable indication of an immunologically hot phenotype. Dr. Brad Nelson at BC Cancer’s The Deeley Research Centre in Victoria, has developed multiplex immunohistochemistry panels to evaluate the tumour microenvironment. Experiments performed in his lab on the sample from the first patient supported the RNA-based results, and confirmed the presence of cytotoxic CD8+ T cells, plasma cells and PD-L1 expressing tumour cells.
Armed with this confirmation, we turned our attention to understanding whether the dominant T cell sequence identified from the RNA data represented a T cell clone that recognized tumour cells. Given the absence of a single clonal mutation, we hypothesized that brachyury, a protein that is typically only expressed in the developing embryo, may have been recognized by the dominant T cell clone.
Our colleague from Canada’s Michael Smith Genome Sciences Centre (GSC) at BC Cancer, Dr. Robert Holt, and members of his lab, tested this hypothesis using an in vitro assay. In these experiments, antigen presenting cells that expressed the patients class I HLA alleles identified from POG sequencing data were either co-expressed with full length brachyury, or incubated with putative brachyury peptide antigens based in silico predictions. Brachyury antigen presenting cells were co-cultured with T cells expressing the dominant T cell clones identified from POG sequencing and confirmed the dominant tumour-associated T cell clone was indeed activated by brachury.
Are these observations generalizable to other tumour types? Overall, trials of immune checkpoint inhibitors in pediatric solid tumours have failed to demonstrate broad response in unselected populations; however, emerging reports have pointed to specific subtypes of pediatric and young adult cancers that may respond. In particular, a subset of tumours with alterations in SWI/SNF complex genes and the presence of immune cell infiltration have demonstrated responses, including SMARCB1-deficient rhabdoid tumours.4–5
Previous research from the Marra lab3 has shown that a subset of rhabdoid tumours characterized by distinct methylation profiles are immunologically hot. Dr. Martin Hirst at the GSC profiled both pediatric POG chordomas using whole genome bisulfite sequencing and methylation profiles along with methylation array datasets from Dr. Marcel Kool at the German Cancer Research Center were clustered with previously published rhabdoid tumour methylation array data.3 Pediatric chordomas clustered most closely with a subgroup of rhabdoid tumours that also had high levels of tumour-associated CD8+ T cells.
Given similarities in the molecular profiles of chordoma and this subgroup of immunologically hot rhabdoid tumours, we propose that a subset of rare, difficult to treat pediatric malignancies may respond to immune checkpoint inhibitor therapy.
Future implications
The observations presented in our article add to previous reports that a rare subset of pediatric malignancies may respond to immune checkpoint inhibitor therapy. Interestingly, these rare responders may not have high tumour mutation burden, which is an established predictor of response in adult cancers. Immunological and genomic profiling will be important for determining which patients may respond to therapy. Establishment of prospective trials that include immune profiling as part of patient selection are required to evaluate the predictive value of these features for patient response.
Personalized OncoGenomics (POG)
The Personalized OncoGenomics (POG) program at BC Cancer, Vancouver, Canada, is led by the directors of Canada’s Michael Smith Genome Sciences Centre (GSC) at the BC Cancer Research Institute, Drs. Marco Marra and Steven Jones and a medical oncologist at BC Cancer, Dr. Janessa Laskin. The study is built on a foundation of collaboration between medical oncologists, pathologists, bioinformaticians, scientists and the broader research community. Pediatric and young adults with hard to treat cancers have a significant unmet clinical need and were included early on in the POG program. Drs. Rebecca Deyell and Rod Rassekh, pediatric oncologists at BC Children’s Hospital, lead the pediatric arm of POG.
Chordoma
Chordoma is a slow growing cancer of spinal tissue, most often found near the tailbone or where the spine meets the skull. They develop from notochord cells that are important in the development of the spinal cord but that do not clear after birth. Most people are diagnosed with chordoma in their 50s and 60s, though pediatric cases make up five per cent of all chordoma diagnoses.
https://www.cancer.gov/pediatric-adult-rare-tumor/rare-tumors/rare-bone-tumors/chordoma
https://www.chordomafoundation.org/
References
- Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12, 453–457 (2015).
- Bolotin, D. A. et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 12, 380–381 (2015).
- 3. Chun, H.-J. E. et al. Identification and Analyses of Extra-Cranial and Cranial Rhabdoid Tumor Molecular Subgroups Reveal Tumors with Cytotoxic T Cell Infiltration. Cell Rep 29, 2338-2354.e7 (2019).
-
Bourdeaut, F., Thaku, M. D., Bergthold, G. & Karski, E. ATRT-11. MARKED RESPONSE TO ATEZOLIZUMAB IN A PATIENT WITH RHABDOID TUMOR: A CASE STUDY FROM THE IMATRIX-ATEZOLIZUMAB TRIAL. Neuro-Oncology 19, iv3–iv3 (2017).
-
Forrest, S. J. et al. Genomic and Immunologic Characterization of INI1-Deficient Pediatric Cancers. Clin Cancer Res 26, 2882–2890 (2020).
Follow the Topic
-
npj Precision Oncology

An international, peer-reviewed journal committed to publishing cutting-edge scientific research in all aspects of precision oncology from basic science to translational applications to clinical medicine.
Related Collections
With Collections, you can get published faster and increase your visibility.
AI Approaches in Drug Design
Publishing Model: Open Access
Deadline: Mar 31, 2026
Genomic Instability
Publishing Model: Open Access
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in