Plasmonic Scattering Imaging of Single Proteins and Binding Kinetics
Published in Protocols & Methods
Written by Pengfei Zhang and Shaopeng Wang
Single molecule imaging techniques permit analysis of protein heterogeneity, measurement of intrinsic properties of individual molecules, and study of molecular binding processes at a level of detail in addition to ensemble approach, which can only describe average expression of multi individual molecules.
Surface plasmon resonance (SPR) is an indispensable tool for detecting molecules and quantifying their binding kinetics without labels. Our idea of detecting single proteins with SPR was first floated back to the year 2000, between Nongjian (NJ) Tao and Shaopeng Wang, then a postdoc at NJ’s lab at Florida International University. We were working on surface plasmon resonance spectroscopy detection of a monolayer of molecules. Our calculation shows that if we can focus the light to a diffraction limited spot, the signal to noise ratio should be sufficient to pick up a single protein binding signal.
We started to work on SPR microscopy (SPRM) in 2008, which offers high spatial resolution, allowing imaging of single cells, sub-cellular organelles, virions, exosomes, nanoparticles, and nanobubbles. However, the extension of SPR to the detection of single proteins is challenging, because the scattering cross sections of proteins are expected to be about million times smaller than those of virions, and the detection is easily affected by the intensity fluctuations of light source and mechanical drifts of system components.
We overcome these limitations by imaging plasmonic waves scattered by the proteins with a second objective placed on top of the sample, in addition to recording the traditional SPRM images from the bottom (Figure 1a). To differentiate this plasmonic imaging method from SPRM, we refer it to as plasmonic scattering microscopy (PSM).
PSM avoids the collection of the strong reflection, allowing thousands of times higher incident light intensity than typical SPRM, and also eliminates the parabolic tail, providing a high contrast image. The high incident intensity not only provides high signal noise ratio for single protein detection, but also allows short average period to minimize the impact of system drift on the measurement. The high contrast image without parabolic tail significantly decreases the difficulty of data processing, making the commonly used software, such as ImageJ, to be available for automated intensity calculation. Moreover, we take advantage of the plasmonic waves scattered by surface roughness, which is usually considered as a noise source in imaging applications, to generate a reference field for interacting with the plasmonic wave scattered by proteins, which can amplify the expected signal by thousand times via interference effect.
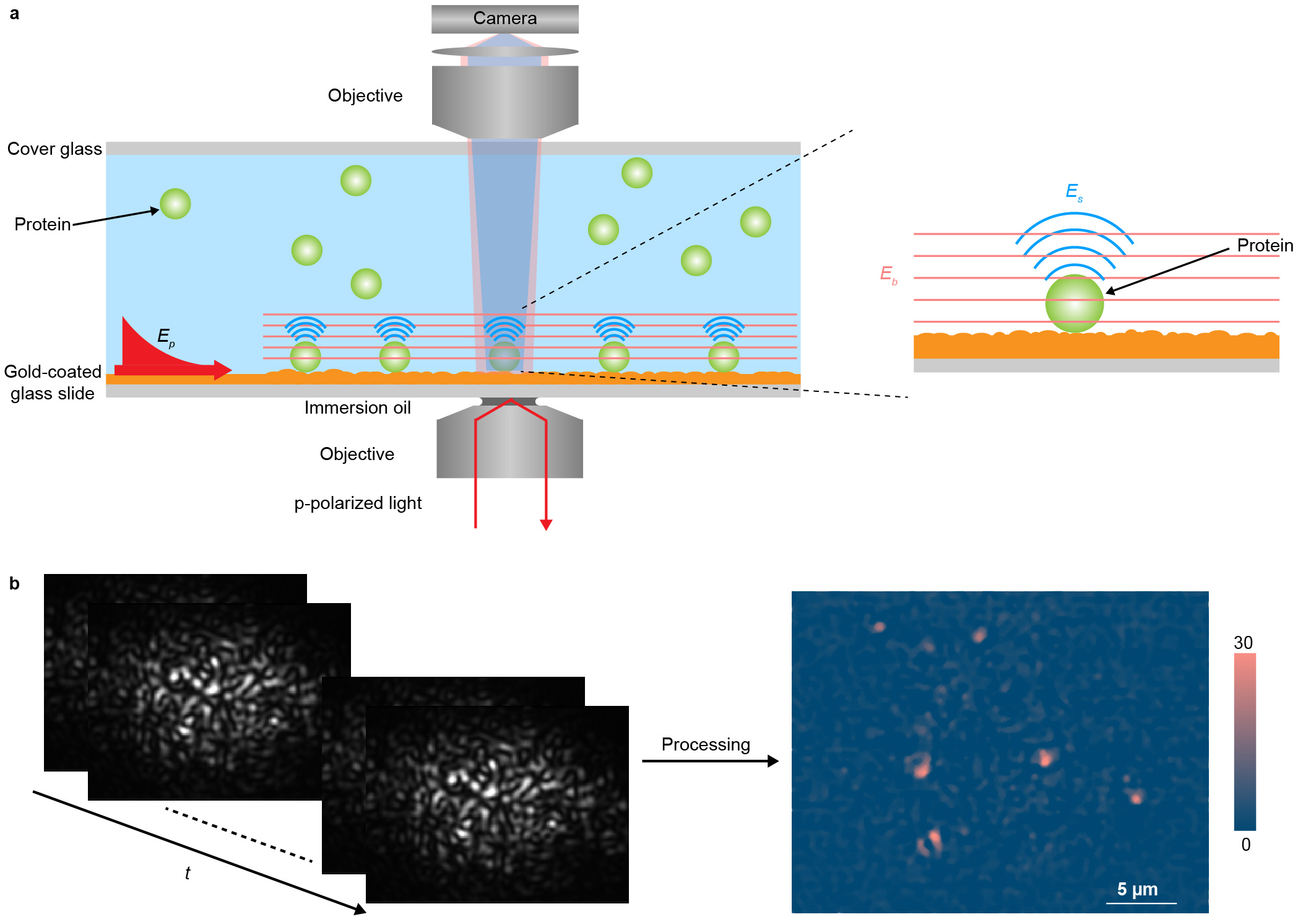
We apply PSM for identification of proteins and measurement of binding kinetics. The experiment shows that the target protein can be bound, and other proteins are just transiently showing up on the antibody modified surface, revealing the specific binding process at single molecule level. In addition to the traditional ensemble approach for binding kinetic measurement, PSM can quantify binding kinetics by digital counting of protein binding events, or by tracking individual protein molecule binding behaviors over time. This single molecule based kinetic measurement can monitor the heterogeneity of protein binding behavior due to conformation, orientation and location on the sensor surface.
Overall, we have demonstrated SPR imaging of single proteins by measuring scattering of plasmonic waves. PSM enables measurement of single proteins and their binding kinetics and is fully compatible with simultaneous traditional SPR measurements. We anticipate that PSM will become a powerful single protein analysis tool for studying various molecular processes.
Prof. NJ Tao unexpected passing away in March 2020, after the initial submission of the manuscript. He was a brilliant scholar and scientist with impactful contributions in the advancement of techniques for nanoscale measurements in areas of molecular electronics, optical imaging and biosensing. A memorial website has been setup for NJ: http://www.njtaoasu.org
Link to the full article: https://www.nature.com/articles/s41592-020-0947-0
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in