
Nucleic acids underpin Darwinian evolution in biology. But where do nucleic acids come from? RNA is the leading candidate for biology’s first replicating molecule, and accordingly its prebiotic synthesis is a holy grail for some in the origins of life research community and has been described as “the molecular biologist’s dream” [1]. However, the strict regio- and stereochemical requirements of furnishing RNA, rather than its many analogues, have frustrated many attempts to elucidate a selective synthesis, such that RNA synthesis is also known as “the prebiotic chemist’s nightmare”! These challenges kindled the idea that another genetic molecule might be easier to make, and the “XNA-world hypothesis” was born – suggesting the first forms of life could have used (non-biological) xenonucleic acids [2]. We were intrigued by xenobiology, but having spent years elucidating prebiotic RNA syntheses [3-9], we were struck that no successful, selective prebiotic XNA syntheses had been published.
We had previously established prebiotic syntheses of anhydro-cytidine [4] and anhydropurines [8] that ostensibly contain the correct stereochemistry to yield arabinonucleic acids (ANAs). We knew anhydro-cytidine hydrolysis afforded ANA nucleosides, arabino-cytidine and arabino-uridine, however the purines, arabino-adenosine and arabino-guanosine, were going to be more challenging. Anhydropurines and anhydro-cytidine can be accessed simultaneously [8], but they need to be manipulated differently to yield the correct nucleobase structures. That was our challenge; what (prebiotic) reaction conditions would simultaneously hydrolyse anhydro-cytidine and reduce anhydro-purines?
Our first clue that this differentiation might not just be feasible but actually predisposed, was that the anhydro-purines simply would not hydrolyse. Then, correlating the principles of site-specific reduction with geochemical analysis, we reasoned sulfide could be the key to purine reduction. Remarkably, we found that H2S (a product of volcanic outgassing) selectively positioned sulfur at the required reduction site, and subsequent UV-irradiation afforded adenosine arabinoside in high yield. Everything was proceeding to plan. We had uncovered a geochemically plausible strategy to discriminate anhydro-cytidine and anhydro-adenosine, which afforded their respective arabinosides in excellent yield [10].
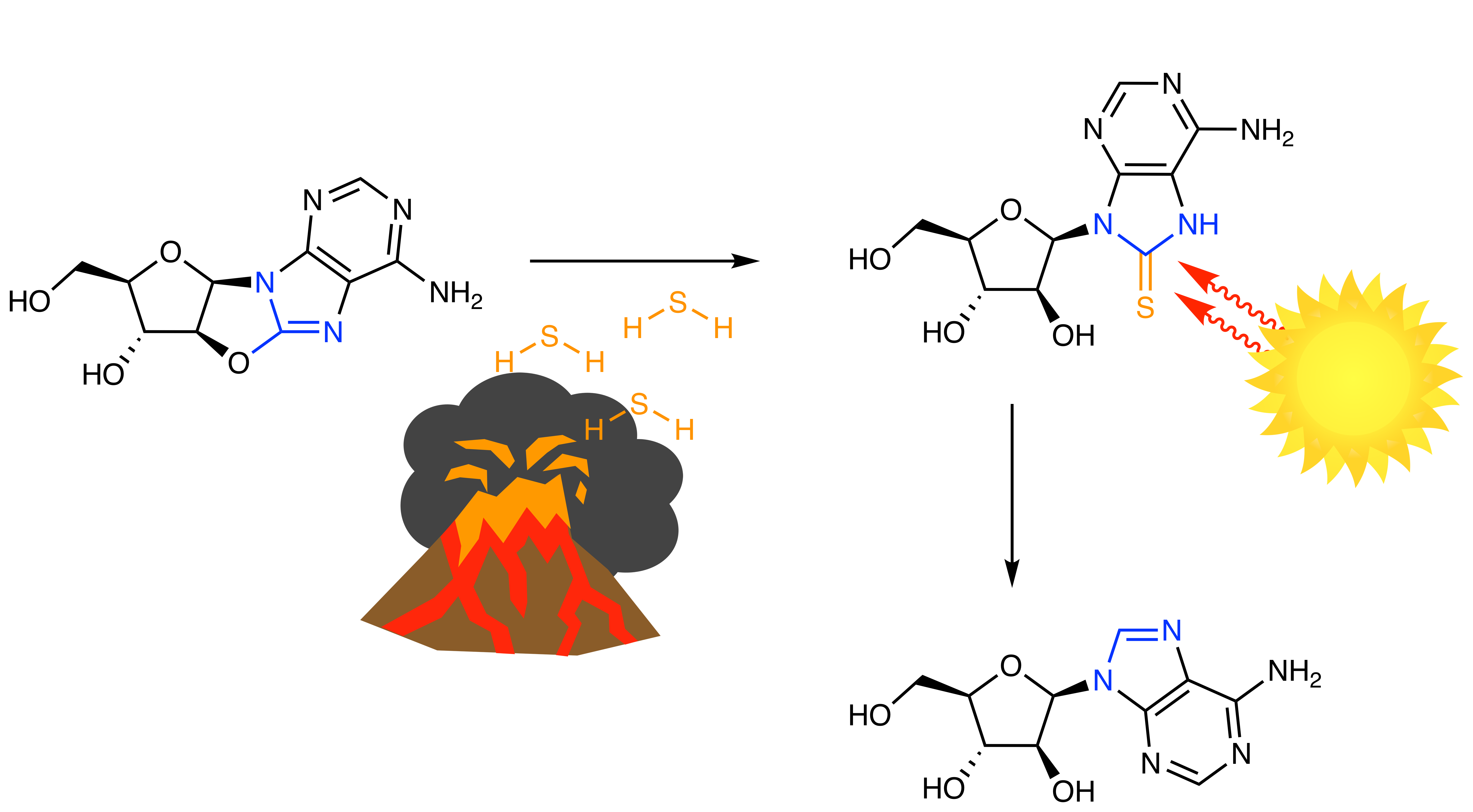
So we next turned our attention to the other purines. Reduction of anhydro-guanosine proceeded just as efficiently as anhydro-adenosine (if not slightly better), but then to our surprise (and delight) we observed irradiation of non-canonical mercapto-inosine gave comparatively poor yields [10]. Selective reduction of mercapto-purines was not dependent on the arabino-stereochemistry, and was equally effective in the natural ribo-stereochemistry [10]. Unlooked for we had found a mechanism to selectively furnish the canonical purines.
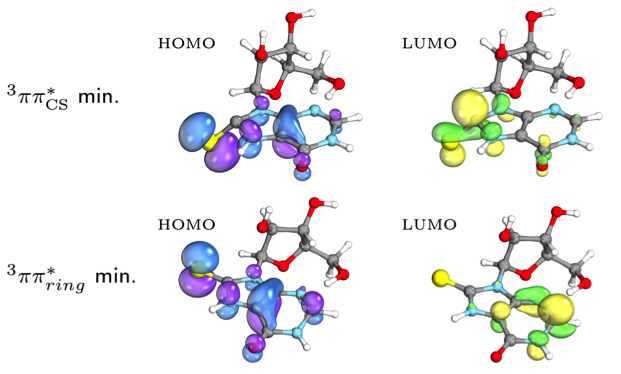
To explain this intriguing selectivity, we turned to our collaborators. Dr Rafal Szabla (Polish Academy of Sciences) revealed, by quantum chemical calculations, subtle differences between photo-excited states of these mercapto-purines, including an unusually low energy and long-lived triplet excited state (3ππring*) enabling degradation pathways in inosine, over C8-reduction (Fig. 2) [10]. Prof. Dimitar Sasselov and Zoe Todd (Harvard-Smithsonian Center for Astrophysics) then measured and analysed the transient absorption spectra of our mercapto-purines, and pleasingly found a good match with the theoretical results, validating our hypotheses.
ANA has demonstrated catalytic prowess and the capability to communicate information with DNA and RNA [11][12]; but did ANA play a pivotal role in life evolution? Or ultimately, if we can combine selective purine-reduction [10] with C2'-stereochemical inversion to afford the ribo-stereochemistry [8], will a complete selective prebiotic synthesis of RNA be the final reward for studying ANA synthesis?
This research has been published in Nature Communications - please click here to read more.
Written by Samuel J. Roberts and Matthew W. Powner, University College London, UK.
References
[1] Orgel, L. E. Crit. Rev. Biochem. Mol. Biol. 39, 99–123 (2004).
[2] Joyce, G. F., Schwartz, A. W., Miller, S. L. & Orgel, L. E. Proc. Natl. Acad. Sci. U.S.A. 84, 4398–402 (1987).
[3] Powner, M. W., Gerland, B. & Sutherland, J. D. Nature 459, 239–242 (2009).
[4] Powner, M. W., Sutherland, J. D. & Szostak, J. W. J. Am. Chem. Soc. 132, 16677–16688 (2010).
[5] Bowler F. R. et al. Nat. Chem. 5, 383-389 (2013).
[6] Engelhart, A. E., Powner M. W. & Szostak J. W. Nat. Chem. 5, 390-394 (2013).
[7] Islam S., Bucar D.-K. & Powner M. W. Nat. Chem. 9, 584–589 (2017)
[8] Stairs, S. et al. Nat. Commun. 8, 15270 (2017).
[9] Fernandez Garcia C., Grefenstette N. M. & Powner M.W. Chem. Commun. 54, 4850–4853 (2018)
[10] Roberts et al. Nat. Commun. 9, 4073 (2018).
[11] Pinheiro, V. B. et al. Science 336, 341–344 (2012).
[12] Taylor, A. I. et al. Nature 518, 427–430 (2015).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in