Precisely programming gene expression in plants
Published in Bioengineering & Biotechnology, Cell & Molecular Biology, and Plant Science
Explore the Research

CRISPRi-based circuits to control gene expression in plants - Nature Biotechnology
Programmable and reversible CRISPRi-based genetic circuits function in a variety of plants.
Background
Gene expression can be programmed using synthetic gene circuits, which are analogous to circuits found in electronic devices but are assembled from interacting biological parts such as DNA, RNA, and proteins. These gene circuits enable cells to sense and respond to various intrinsic and extrinsic signals. This enables different activities of promoters to drive gene expression only under certain input combinations. However, the development of such gene circuits in plants have been hindered by the limited availability of modular biological parts required for their construction. Therefore, there is a need to construct more sophisticated synthetic gene circuits that would enable us to dictate the exact timing (WHEN) and cell type (WHERE) a particular gene is switched ON or OFF.
The inspiration
In 2014, inspired by the synthetic gene circuit technology advances in microorganisms and mammalian cell lines1–3, we aimed to create a more sophisticated gene circuit platform to control gene expression in plants. At the time, different tools were used for the construction of gene circuits that included DNA binding proteins, recombinase enzymes, and CRISPR interference (CRISPRi).
To develop synthetic gene circuits in plants, we chose to use CRISPRi that employs nuclease deactivated Cas9 (dCas9) enzyme for targeted gene repression. Synthetic gene circuits based on CRISPRi are modular, reversible and can easily be reprogrammed by changing single guide RNAs (sgRNAs), which are used as inputs.
While CRISPRi had been demonstrated for gene regulation in plants, its effectiveness in developing reversible gene circuits was still unknown. We were keen to explore the potential of CRISPRi in the development of synthetic gene circuits, aiming to advance programming gene expression in plants.
The Struggle
To turn our ideas into reality, we initiated our project in 2015 to create reversible synthetic gene circuits in plants. However, establishing a new research area in the lab presented significant challenges. Initially, we focussed on two key aspects. First, we aimed to develop a high-throughput assay for screening genetic parts and circuits given the long life cycles of plants. Second, we set out to create a CRISPRi-based NOR gate, often referred to as a “universal gate” that can be layered to perform complex logic operations.
To develop a high-throughput and robust screening assay for testing the performance of gene circuits, we optimized a 96-well plate-based protoplast transfection and dual luciferase assay using Arabidopsis leaf protoplasts. However, this optimization took us a while, as we were facing high variation in the assay results. One possible reason for this could be the transfer of transfected cells to a new plate containing the buffer for incubation. This step typically required centrifugation of the transfected cells to be collected at the bottom. However, one day, while performing the experiment, the centrifuge stopped working! As we had spent the whole day getting to the last step, we were unsure what to do. After 45 mins, we noticed that the cells were settled down at the bottom, likely simply due to gravity, and could be easily collected after removing the supernatant. However, instead of moving the cells to a new plate, we added the buffer required for incubation to the same plate, and that was the moment when we started getting reproducible data. This suggested that by moving the transfected cells to a new plate, we used to lose transfected cells that were causing variation in our data. This was the turning point in the project, and the magic started to happen. However, it was just the beginning as the next challenge was identification of the regions for enhanced CRISPRi in plant promoters. By engineering different regions in the promoter, we figured out that targeting regions immediately upstream and downstream of the TATA box results in enhanced CRISPRi-based repression. We referred to such engineered promoters as “integrators” for their ability to receive and integrate input signals in the form of sgRNAs to control the output. It took us 3 years to implement a CRISPRi-based NOR gate in plant cells (Fig. 1)!
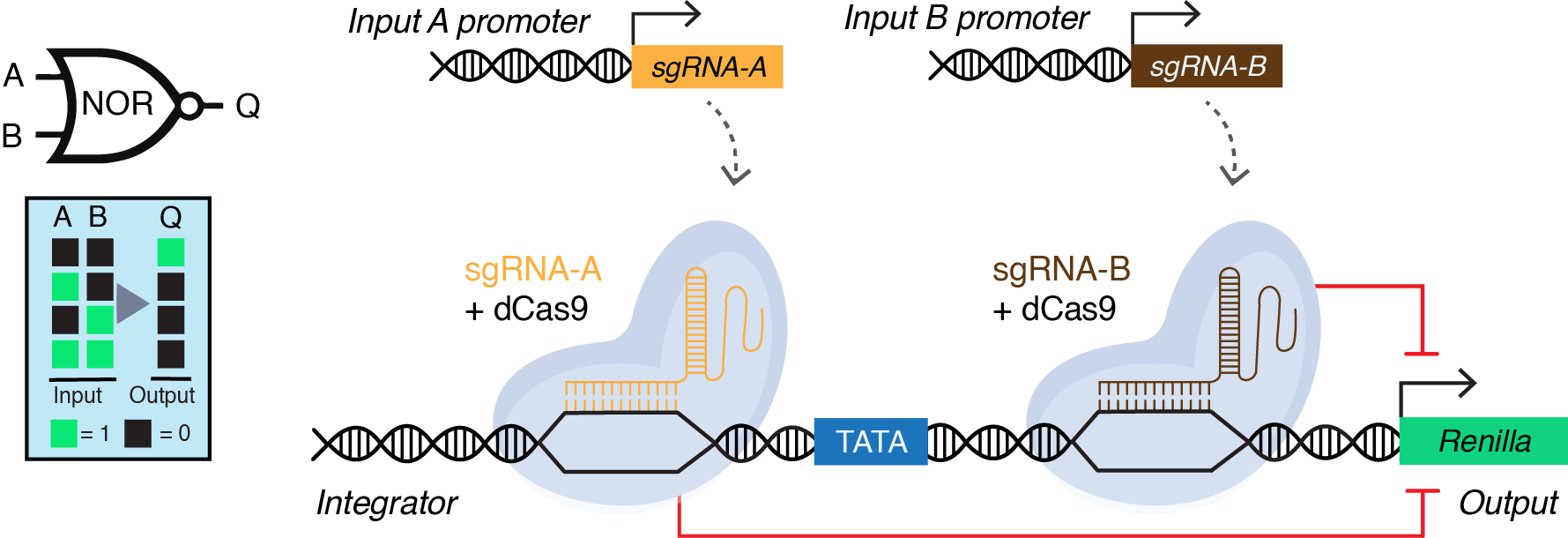
The successful implementation of a NOR gate in plant cells gave us a new hope that we could build on this system. However, the next challenge was achieving programmability of the NOR gate for spatio-temporal control of gene expression. For this purpose, we had to test three different systems: ribozymes, tRNAs, and the Csy4 endoribonuclease system to process the input sgRNAs from a Pol-II promoter. The next and most critical challenge was the implementation of a CRISPRi-based NOR gate in stably transformed plants to demonstrate programmability and reversibility. For this purpose, we used heat and dexamethasone (DEX) inducible promoters for expressing the input sgRNA-A and sgRNA-B, respectively. However, here in the presence of heat-induced input sgRNA-A the integrator promoter was repressed, however, the DEX-induced input sgRNA-B was unable to repress the integrator. By doing a thorough analysis we figured out that the heat-inducible promoter was 5x stronger than the DEX-inducible promoter, and therefore, less input sgRNA-B was produced and it was insufficient to switch off the integrator. We confirmed this finding by testing different strength input promoters on their ability to repress our designed integrators. This revealed that the amount of sgRNA production is an important design factor in building CRISPRi-based circuits in plants. Long story short! With the help of a great team, we were able to add to the recently developed memory4 and transcriptional5 plant synthetic circuit systems and develop a CRISPRi-based gene circuit platform that is compact, modular, programmable, and reversible. This was an exciting journey of almost 9 years! We are excited to apply these circuits in our recently established ARC Centre of Excellence in Plants for Space to reprogram plants for space as well as challenging or controlled environment conditions on earth.
References
- Gaber, R. et al. Designable DNA-binding domains enable construction of logic circuits in mammalian cells. Nat. Chem. Biol. 10, 203–208 (2014).
- Kiani, S. et al. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 11, 723–726 (2014).
- Nielsen, A. A. K. & Voigt, C. A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 10, 763 (2014).
- Lloyd, J. P. B. et al. Synthetic memory circuits for stable cell reprogramming in plants. Nat. Biotechnol. (2022) doi:10.1038/s41587-022-01383-2.
- Brophy, J. A. N. et al. Synthetic genetic circuits as a means of reprogramming plant roots. Science 377, 747–751 (2022).
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in