Real-time disease monitoring predicts early relapse and refines conventional imaging in patients with DLBCL
Published in Cancer, Genetics & Genomics, and Biomedical Research
The challenge of monitoring treatment in lymphoma
Diffuse large B-cell lymphoma (DLBCL) is the most common type of aggressive lymphoma. For decades, frontline therapy has centered on R-CHOP. While recent treatment combinations including novel agents such as tafasitamab and lenalidomide has improved outcomes for many patients, relapse remains common and unpredictable using conventional imgaging.
Positron emission tomography (PET-CT) has been the gold standard for assessing treatment response within the last decades, however, its sensitivity and specificity are limited. False positive results can lead to anxiety, unnecessary biopsies, and delays in decision-making. This limitation has spurred the search for better biomarkers.
For B-cell lymphomas, malignant cells carry unique immunoglobulin (IG) gene rearrangements, which serve as patient-specific molecular fingerprints. Using next-generation sequencing (NGS), these rearrangements can be identified and tracked over time.
Cell free (cf)DNA consists of DNA fragments shed by normal or cancer cells into the bloodstream and can be isolated from plasma. cfDNA molecules of tumor origin are often called circulating tumor (ct)DNA. Longitudinal ctDNA assessment can detect minimal residual disease (MRD) with high sensitivity, track treatment kinetics in real time, and potentially refine or even replace certain imaging-based assessments.
Our study: embedding ctDNA analysis in a clinical trial
The FirstMIND trial was designed to evaluate the feasibility of tafasitamab plus R-CHOP, with or without lenalidomide, in newly diagnosed DLBCL. Beyond clinical outcomes, we embedded a prospective ctDNA analysis using the EuroClonality IG-NGS assay. Our aims were threefold:
-
Feasibility: Can IG clonotypes be reliably detected from plasma without depending on tumor biopsies?
-
Clinical utility: Can ctDNA detection predict treatment outcomes?
-
Complementarity: Can ctDNA refine PET findings and reduce uncertainty in response assessment?
Feasibility: clonotype detection from plasma
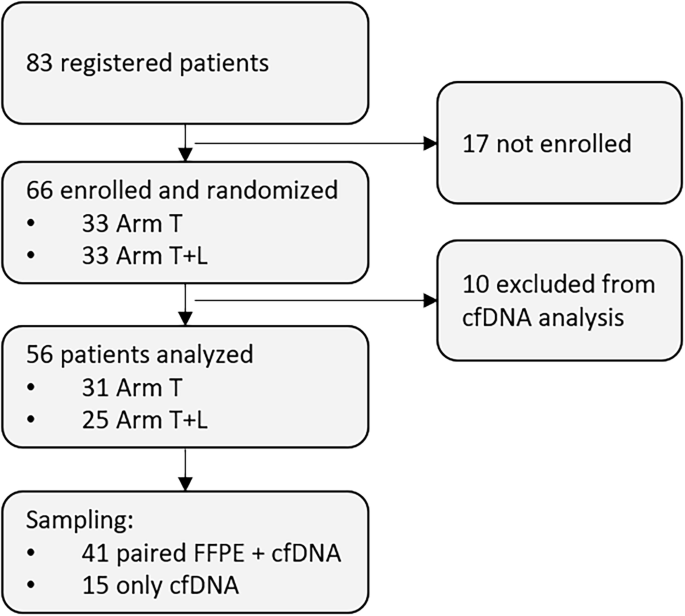
We analyzed more than 300 samples from 56 patients. Disease-related clonotypes were identified in 89% of cases, and in the majority, plasma cfDNA alone was sufficient. This is clinically meaningful: it demonstrates that plasma cfDNA offers access to tumor material without the need of invasive tumor biopsies.
By validating clonotypes against matched tumor tissue when available, we confirmed the robustness of this liquid biopsy-based approach. While a small fraction of patients lacked detectable clonotypes due to polyclonal signals, the overall success rate supports the use of ctDNA testing in real-world frontline practice.
Tracking treatment response in real time and complementing PET-CT
Once clonotypes were identified, we tracked ctDNA at predefined time points: after cycle 1 (C2D1), after cycle 3 (C4D1), at end of treatment (EOT), and during follow-up. By C2D1, 52% of patients were already ctDNA-negative. By C4D1, that figure rose to 83%, and by EOT, 82% remained negative. Six months later, 93% of patients had sustained ctDNA clearance.
These dynamics were not only impressive but also prognostic. Patients who remained ctDNA-positive after early cycles had significantly worse progression-free survival (PFS) and overall survival (OS). For example, ctDNA positivity at C2D1 translated to a hazard ratio of 4.51 for PFS. At EOT, ctDNA positivity predicted a six-fold higher risk of progression. This highlights the unique value of ctDNA: it identifies high-risk patients much earlier than clinical or imaging endpoints, opening the door to adaptive interventions.
While PET remains central in lymphoma, its imperfections are currently under debate. In our study, ctDNA proved particularly helpful in clarifying PET results. Among patients who were PET-positive at the end of treatment, five were ctDNA-negative. None of these patients relapsed during follow-up. in contrast, all PET-positive ctDNA-positive patients relapsed within six months.
This suggests that ctDNA can distinguish true residual disease from false-positive PET signals, improving specificity. Such integration could spare patients unnecessary biopsies and guide clinicians with greater confidence.
cfDNA offers more than on-treatment monitoring
Interestingly, ctDNA was also informative at diagnosis. Baseline cfDNA and ctDNA levels correlated with established prognostic factors such as elevated LDH and higher International Prognostic Index (IPI) scores. Patients with high cfDNA levels (≥3.35 log10 hGE/ml) had significantly worse outcomes, confirming the prognostic role of ctDNA even before treatment begins. This supports the idea that ctDNA can refine risk stratification, helping clinicians identify high-risk patients who might benefit from more intensive regimens or clinical trial enrollment.
Technical insights and challenges
While encouraging, our study also shed light on technical considerations. IG-NGS proved reliable and widely applicable, but its sensitivity is lower than other mutation-based approaches such as CAPP-Seq or PhasED-Seq. Differences in assay design, input requirements, and thresholds remain barriers to standardization.
Another limitation is biological: ctDNA may fail to capture disease in sanctuary sites. For instance, one patient who was ctDNA-negative at EOT later relapsed with central nervous system involvement. This underscores the need for complementary modalities, recognizing both the strengths and boundaries of ctDNA.
Looking forward: ctDNA in adaptive therapy
The broader vision is clear: ctDNA could guide adaptive therapy. Patients who remain ctDNA-positive after initial cycles could receive intensified treatment to prevent relapse, while those who clear ctDNA rapidly might be candidates for shorter or less intensive therapy.
This approach would personalize treatment based on real-time molecular response rather than fixed schedules or imaging alone.
Reflections
For us, this project was about redefining how to monitor lymphoma. PET-CT will remain essential, but ctDNA provides a new layer of precision. It can capture the hidden battle at the molecular level, offering clinicians and patients earlier, more reliable insights into treatment success or failure.
Our findings show that ctDNA is not only feasible in the frontline setting but also clinically meaningful. It can refine prognostication, monitor response in real time, and clarify PET results. With further validation and standardization, we believe ctDNA will soon become an integral part of routine lymphoma care, helping us move toward a future of more personalized, adaptive treatment.
Follow the Topic
-
Leukemia

This journal publishes high quality, peer reviewed research that covers all aspects of the research and treatment of leukemia and allied diseases. Topics of interest include oncogenes, growth factors, stem cells, leukemia genomics, cell cycle, signal transduction and molecular targets for therapy.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in