REDit: Engineering erythroid cells for therapeutic applications
Published in Bioengineering & Biotechnology

Genome editing technologies are rapidly changing our approach to treat genetic disorders. RNA-guided nucleases such as Crispr/Cas9 and base editors can potentially fix most disease-causing mutations, intervening directly on the patient’s genome1,2. However, the development of endless mutation-specific editing strategies can be a major hurdle for clinical translation, so the idea of developing a versatile integration platform to tackle different genetic disorders with one editing strategy is quite appealing.
Previous studies identified the human albumin gene and its promoter as an ideal locus to achieve high liver-driven expression of integrated transgenes3 and resulted in the first clinical trial in humans using nucleases in vivo (NCT03041324). During my PhD I had the chance to spend long coffee breaks and have great discussions with two of the amazing scientists -Dr. Xavier Anguela and Dr. Rajiv Sharma- who were developing this idea and my fascination for genome engineering began.
Although the approach was promising and exciting, it faced some drawbacks: i) it relied on continuous expression of nucleases that can trigger immune responses4 and increase editing at off-target sites; ii) it depended on in vivo delivery of AAV vectors and therefore is limited by pre-existing (and frequent) antibodies to the viral capsid5; iii) it was liver-specific, a potential issue for genetic disorders with liver damage.
An alternative strategy started taking shape during my postdoc in Dr. Amendola’s lab at Genethon working on genome editing of hematopoietic stem cells (HSC) with CRISPR/Cas9. Differently from liver, HSCs can be easily accessed, edited ex vivo via transient expression of Cas96, and re-administered; however, a suitable locus for transgene integration was yet to be identified.
To maximize protein expression, we had to find a highly transcribed – but somewhat disposable- gene that was present in an abundant hematopoietic cell lineage. α-globin genes were the answer, as they are highly expressed (~1.5g/day) by erythroid cells, and the loss of up to 3 out of 4 α-globin alleles is mostly asymptomatic. As a single erythroid progenitor can produce more than 106 erythroblasts, we hypothesized that the combination of robust transcription and the abundance of transgene-expressing cells would boost protein production, even with few integrations in HSCs.
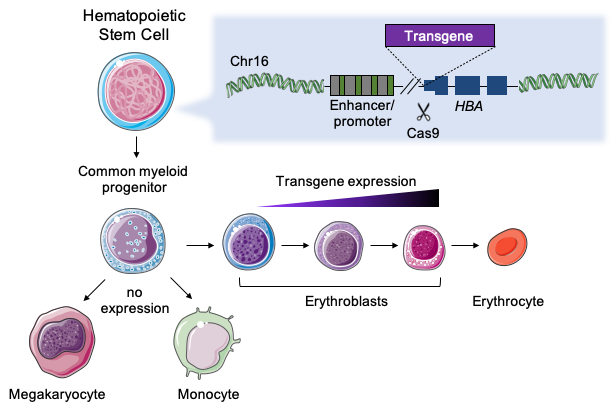
We designed several guide RNA in the α-globin locus to target the insertion and used AAV as DNA template to deliver transgenes involved in different genetic disorders. When choosing the best place to integrate, we had to preserve erythroid-specific regulation and be careful not to interfere with erythropoiesis; the help of Dr. Annarita Miccio was instrumental to achieve these goals. Targeting was extremely efficient in HSCs (>50%) therefore no selection of edited cells was required. With our integration strategy, erythroblasts expressed and secreted functional enzymes at high levels including lysosomal acid lipase (involved in Wolman disease), Factor IX (hemophilia B), α-l-iduronidase (MPSI) and α-galactosidase (Fabry disease).
We believe that this platform could be useful for a number of therapeutic applications, in particular for monogenic disorders that require protein replacement therapy, but also to engineer red blood cells with novel functions to target cancer or to induce self-antigen tolerance in auto-immune diseases.
Our paper:
Pavani, G., Laurent, M., Fabiano, A. et al. Ex vivo editing of human hematopoietic stem cells for erythroid expression of therapeutic proteins. Nat Commun 11, 3778 (2020).
https://doi.org/10.1038/s41467-020-17552-3
References
- Li, H. et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475, 217–221 (2011).
- Rees, H.A. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 19(12), 770-788 (2018).
- Sharma, R. et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 126, 1777–1784 (2015).
- Charlesworth, C. T. et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 25, 249–254 (2019).
- Pien, G. C. et al. Capsid antigen presentation flags human hepatocytes for destruction after transduction by adeno-associated viral vectors. J. Clin. Invest 119, 1688–1695 (2009).
- Lattanzi, A. et al. Optimization of CRISPR/Cas9 delivery to human hematopoietic stem and progenitor cells for therapeutic genomic rearrangements. Mol. Ther. 27, 137–150 (2019).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in