Reproducible Neuroimaging for Everyone – behind the Scenes of Neurodesk
Published in Healthcare & Nursing, Neuroscience, and Computational Sciences
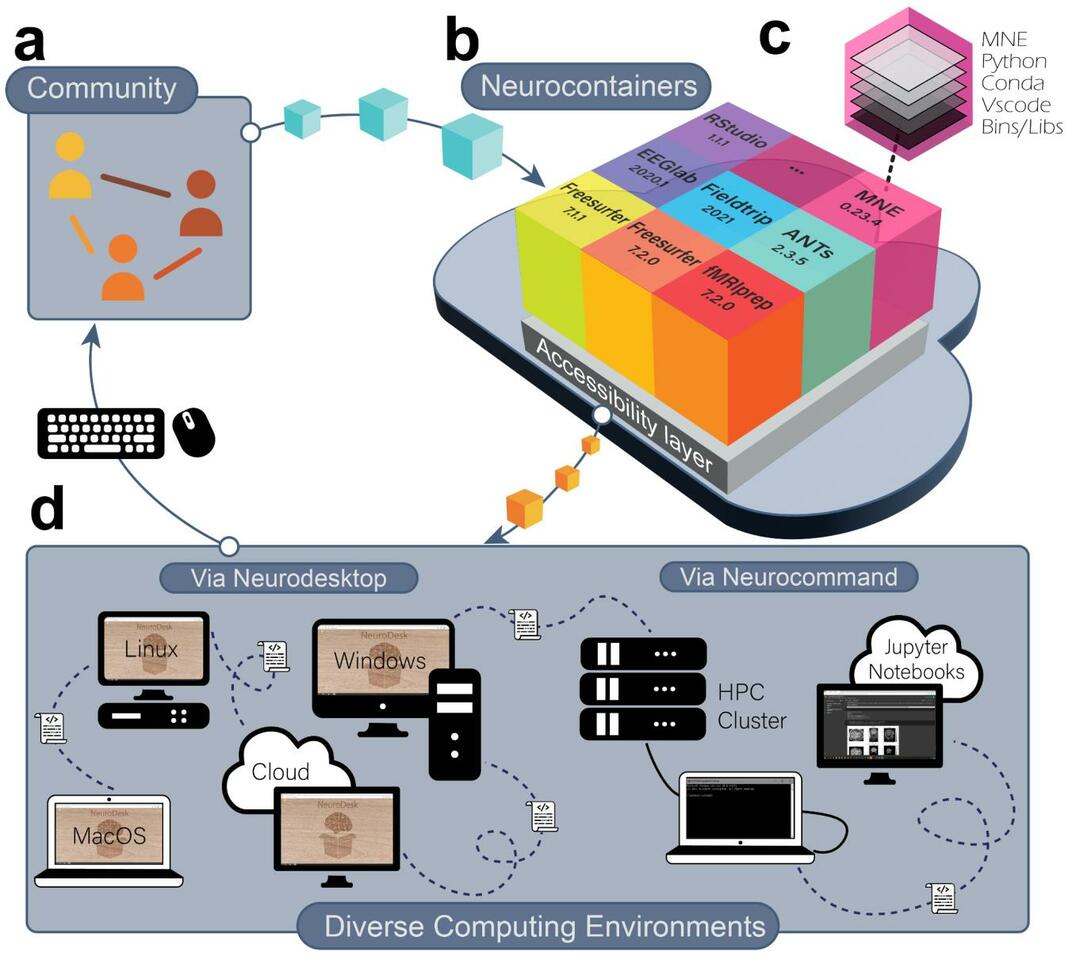
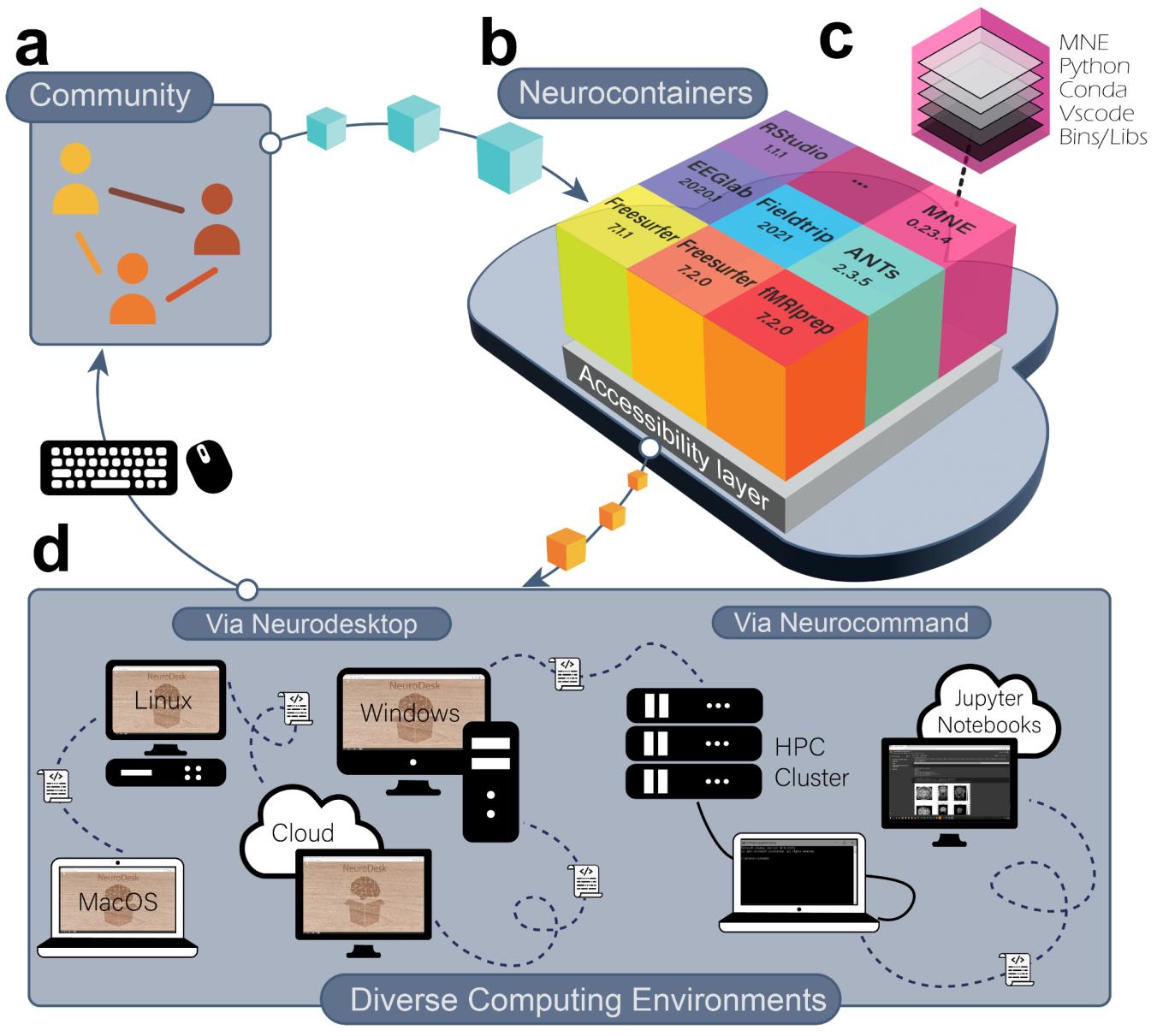
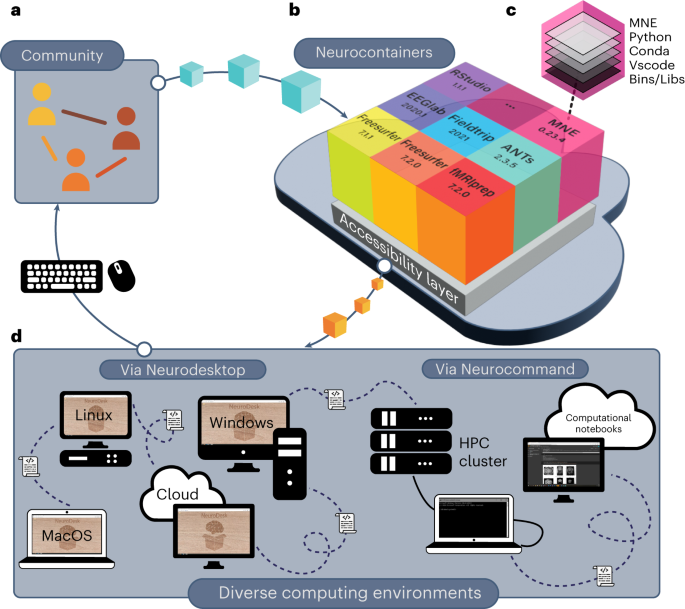
A One-Stop Solution for Neuroimaging Analysis
The development of Neurodesk started with an OHBM Brainhack Hackathon project in 2020 proposed by Oren Civier (https://github.com/ohbm/hackathon2020/issues/177). In this project, the goal was to develop a software container that includes common neuroimaging applications. We created a virtual desktop environment in a software container in just three days. This environment could access different software containers with neuroimaging applications. A prototype was born and named “Neurodesk” - a NEUROimaging DESKtop environment. In the following months, a community collaboration spearheaded by groups at three Australian universities with support from the Australian National Imaging Facility sought funding from the Australian Research Data Commons (ARDC) through their platform project scheme. The project was funded to build the Australian Electrophysiology Data Analytics PlaTform (AEDAPT), with the goal of creating a national platform for reproducible electrophysiology data analysis and sharing, accessible to all Australian researchers across a wide range of disciplines that conduct electrophysiological research. The development of Neurodesk fits this overarching goal of the AEDAPT project and provides a powerful platform to enable seamless reproducible neuroscience.
We believe Neurodesk is a game-changer, offering a comprehensive suite of neuroimaging software containers accessible through a user-friendly browser-based interface. Neurodesk runs on personal workstations, high-performance computers, and cloud environments, ensuring accessibility, flexibility, portability, and full reproducibility of neuroimaging analyses across different computing hardware.
Overcoming Technical Challenges
Building this platform involved tackling multiple technical challenges to create a versatile and accessible data analysis environment for reproducible neuroimaging. Let's dive into the key challenges and how we addressed them:
1. Cross-Platform Compatibility
Picture Dr. Smith, a neuroscientist, who spent hours trying to install analysis software on a Windows or Mac computer that was designed for Linux. This common frustration is what Neurodesk elegantly resolves. Neurodesk's primary challenge was ensuring seamless functionality across different operating systems and browsers. Since many neuroimaging tools run on Linux, the project bridged the compatibility gap by adapting and optimizing these tools to work within a software container accessible through a browser. Neurodesk is compatible with Windows, MacOS, GNU/Linux and even supports Apple Silicon hardware through Rosetta 2 emulation.
2. Dependency Management
Neuroimaging tools often have complex dependencies, leading to issues with incomplete dependencies or version mismatches. Neurodesk implemented robust dependency management and isolation of different software packages, ensuring that required components are readily available and configured correctly, reducing the potential for unreproducible setups. We achieve this by packaging every application into an apptainer/singularity container with all dependencies required to run this software. We built a user-friendly way of creating these containers on GitHub using the open-source tool neurodocker and GitHub actions: https://github.com/NeuroDesk/neurocontainers. This makes adding new applications from anyone in the neuroimaging community straightforward.
3. Scalability
Addressing scalability challenges, Neurodesk efficiently allocates and manages computational resources to accommodate a diverse range of research projects, from small-scale to large-scale analyses, from individuals and small labs to large national or international collaborations. Neurodesk supports running analyses on laptops, workstations, cloud computing and even high-performance computing clusters - with this, researchers can utilize the right resources at the right time.
4. User-Friendly Interface
Neurodesk offers an intuitive and user-friendly interface to lower the entry barrier for researchers. We began with a familiar virtual Linux desktop environment catering to most users. However, recognizing the increasing popularity of Jupyter notebooks among neuroscientists, we integrated a full Jupyterhub system. The design facilitates easy access and utilization of neuroimaging tools, eliminating the need for extensive technical expertise.
5. Reproducibility
Ensuring full reproducibility was a fundamental goal. Neurodesk manages dependencies and implements version control and data management practices that enable researchers to recreate their analyses accurately across different computing systems. Importantly, analysis results across different platforms are identical, which is not the case with traditional installation of software.
Extending the Impact to Other Disciplines
The challenge of ensuring reproducibility is not unique to neuroimaging. Neurodesk's solutions can be adapted to address similar challenges in other fields, such as genomics. The user-friendly, browser-based interface of Neurodesk can serve as a model for creating accessible research environments in disciplines where researchers may lack extensive technical expertise but require advanced computational tools.
Conclusion
In conclusion, Neurodesk marks a significant step towards revolutionizing neuroimaging data analysis. Overcoming technical challenges and offering a user-friendly interface sets a precedent for accessible research environments across scientific disciplines, promising a more reproducible and collaborative future in data analysis. The Neurodesk platform is built by and for the scientific community to enable anyone to perform reproducible analyses easily.
Ready to revolutionize your research with hassle-free access to neuroimaging tools? Join the Neurodesk community today by navigating to https://www.neurodesk.org/, trying out Neurodesk on the cloud or your own computer, and taking your first step towards seamless neuroimaging data analysis.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in