Rhizosphere viruses - just how powerful are they in mediating microbial arsenic metabolism?
Published in Microbiology
The rhizosphere, the zone surrounding plant roots, is a hub of intense biological and chemical interactions1. While much attention has been paid to the microbial life in this region2, the role of viruses has largely been overlooked until now. Soil viruses, which outnumber microbes in soil by a factor of ten, impact biogeochemical processes in the soil both directly and indirectly3, by lysing host cells or shaping host metabolism through lysogeny. The rhizosphere's enhanced nutrient flow and microbial activity likely create a more dynamic and intricate environment, fostering stronger interactions between viruses and their host cells4. This interaction is not just incidental; it could be driving significant changes in the microbiota’s evolution and ecology5. This raises an intriguing possibility: could viruses be playing a more central role in shaping rhizosphere processes than previously thought?
Arsenic (As), a toxic metalloid found globally in soils, poses significant environmental risks6. Rice (Oryza sativa L.) uptake of As is influenced by microbe-mediated As redox changes in the rhizosphere, where its roots promote a microbial community that aids in As oxidation through exudates and oxygen release7. This makes the rice rhizosphere an important model for studying As risk reduction8. However, the potential influence of viruses on As biotransformation remains unknown. Given that flooding increases connectivity between soil microaggregates and intensifies microbial interactions9, we hypothesized that rhizosphere conditions might amplify the role of viruses in influencing microbial As metabolism. This question drove our research, aiming to uncover the hidden connections between viruses, microbes, and As transformation in this complex and ever-changing environment.
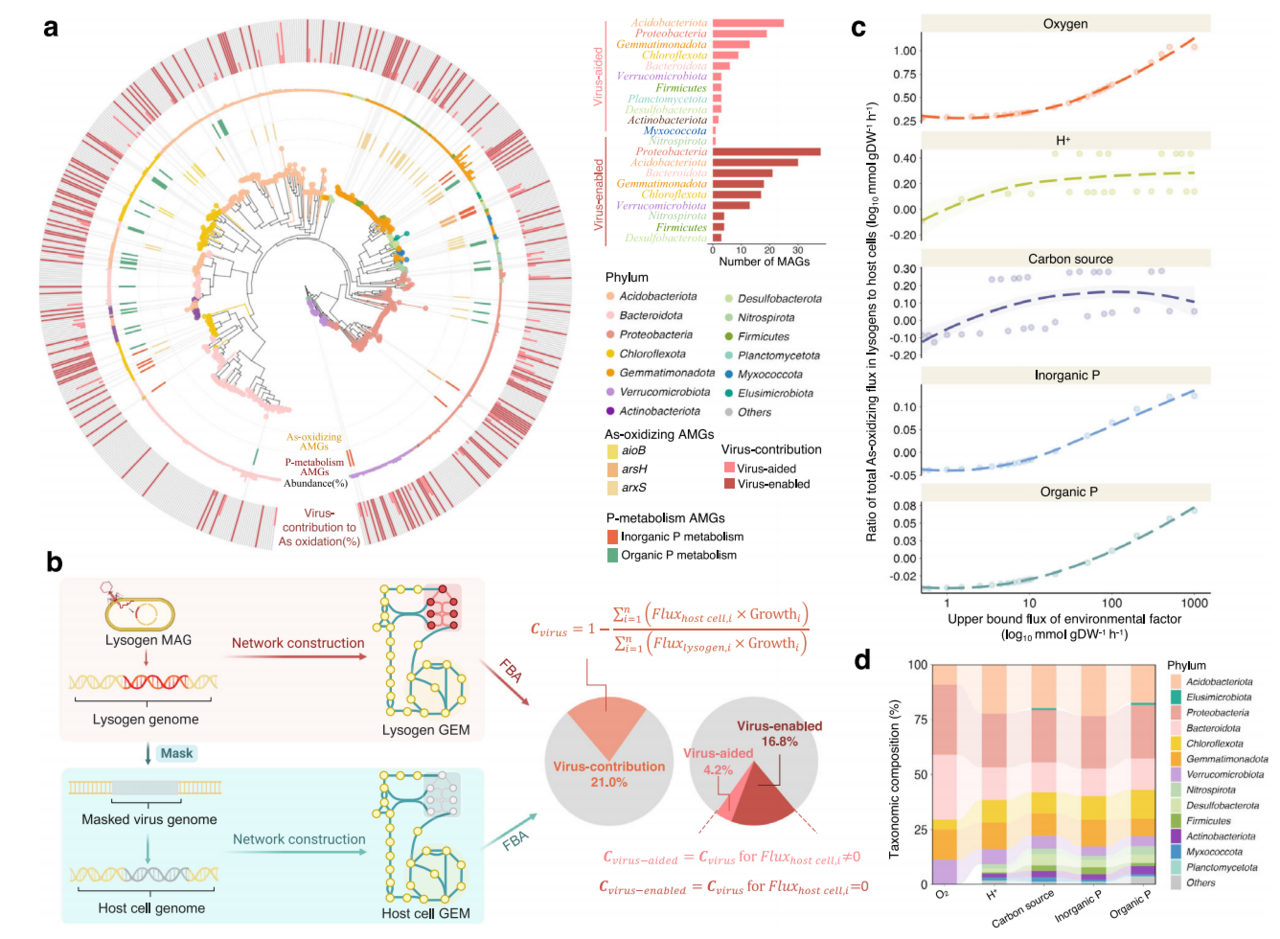
Our study began by exploring the dynamic interactions between viruses and microbes in the rhizosphere. Through monitoring of As species and quantitative time-series metagenomics (Figure 1), we made an unexpected discovery: virus-associated microbes had a far greater influence on As oxidation than the broader microbial community. Most importantly, we found that the rhizosphere triggers lysogeny in viruses associated with As-oxidizing microbes, pointing to a potential mechanism for virus-mediated regulation of microbial activity. Further analysis revealed that rhizosphere lysogenic viruses were enriched with As oxidation and phosphate co-metabolism genes and facilitated horizontal gene transfers (HGTs) of As oxidases.
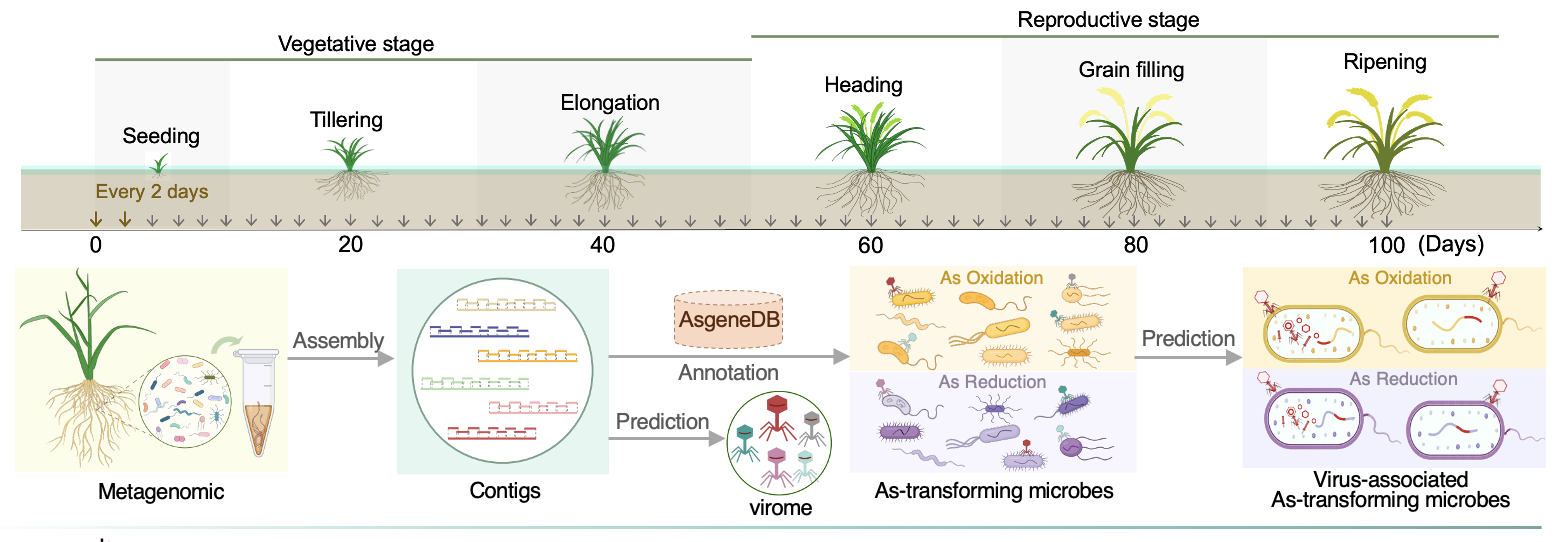
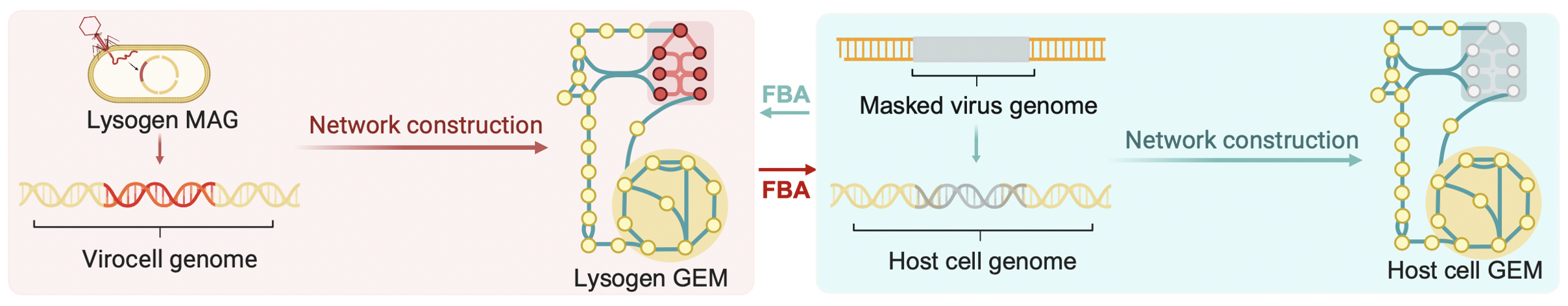
The next big challenge was figuring out how to quantify the interaction between rhizosphere viruses and microbes in driving As oxidation. To tackle this, we turned to genome-scale metabolic models (GEMs) for inspiration (Figure 2). We then developed a pipeline using flux balance analysis (FBA) to focus on As-oxidizing microbes—comparing those with lysogenic viruses (lysogens) and those without (host cells). This in silico comparison allowed us to dive deep into the virus-driven As metabolic network, unraveling the hidden ways viruses might be influencing the process. We also took into account key environmental factors in the rhizosphere, like oxygen, carbon, and phosphorus levels, which helped us understand how these conditions can enhance the role of viruses in As oxidation across different microbial groups.
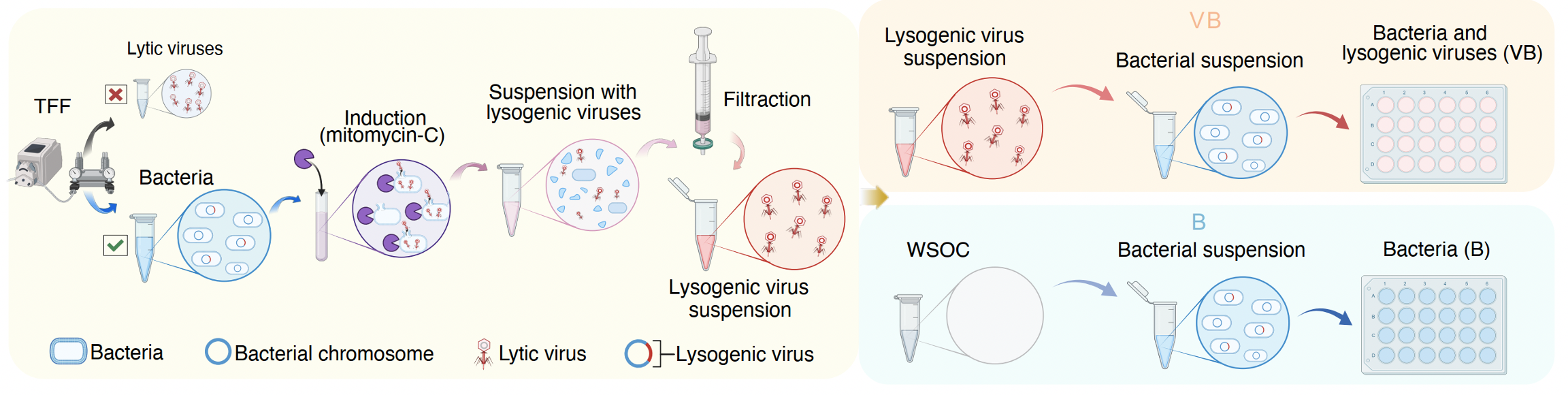
And here is the finally key moment – designing an experiment to validate the contribution of viruses from the rhizosphere and the bulk soil to microbial As transformation (Figure 3). Through in vitro batch culture experiments with induced lysogenic viruses, our results estimated rhizosphere lysogenic viruses contribute to up to 25% of microbial As oxidation. In contrast, the viruses in lysogenic lifestyle from the bulk soil increased the As reduction capacity of microbes.
Our research provides new insights and a framework for exploring plant-microbiome-virus interactions. After nearly five years of work, our findings have been published, marking the conclusion of this journey. To the best of our knowledge, this is the first study to quantitatively evaluate the contribution of soil viruses to rhizosphere processes, highlighting the potential of harnessing rhizosphere viruses to mitigate environmental risks associated with As.
But is this really the end of the journey? With the latest advances in technology and knowledge, we believe that viruses can serve as a powerful predictive tool for rhizosphere dynamics, particularly in the context of rhizosphere biogeochemical cycles. Further investigation into the viral ecological mechanisms related to these cycles will unlock greater potential for leveraging rhizosphere viruses to sustain agricultural soil health.
References
- Korenblum, E. et al. Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. USA 18, 3874–3883 (2020).
- Ling, N., Wang, T. & Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 13, 1–13 (2022).
- Jansson, J. K. & Wu, R. Soil viral diversity, ecology and climate change. Nat. Rev. Microbiol. 21, 296–311 (2023).
- Braga, L. P. P. & Schumacher, R. I. Awaking the dormant virome in the rhizosphere. Mol. Ecol. 32, 2985–2999 (2023).
- Santos-Medellín, C., Blazewicz, S. J., Pett-Ridge, J., Firestone, M. K. & Emerson, J. B. Viral but not bacterial community successional patterns reflect extreme turnover shortly after rewetting dry soils. Nat. Ecol. Evol. 7, 1809–1822 (2023).
- Zhang, S. et al. Escalating arsenic contamination throughout Chinese soils. Nat. Sustain. 1–10 https://doi.org/10.1038/s41893-024-01341-7 (2024).
- Jia, Y., Huang, H., Chen, Z. & Zhu, Y.-G. Arsenic uptake by rice is influenced by microbe-mediated arsenic redox changes in the rhizosphere. Environ. Sci. Technol. 48, 1001–1007 (2014).
- Chen, S.-C. et al. The Great Oxidation Event expanded the genetic repertoire of arsenic metabolism and cycling. Proc. Natl. Acad. Sci. USA 117, 10414–10421 (2020).
- Ma, B. et al. Biogeographic patterns and drivers of soil viromes. Nat. Ecol. Evol. 8, 717–728 (2024).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in