Same driver, different routes: generating a faithful model of MLL-rearranged infant leukemia
Published in Cancer

Siobhan Rice1 and Nicholas T Crump1
1 MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, NIHR Oxford Biomedical Research Centre Haematology Theme, Radcliffe Department of Medicine, University of Oxford, Oxford, UK
Overall, the treatment of childhood acute lymphoblastic leukemia (ALL) has been a remarkable success story, with improved therapies and better supportive care meaning that 9 out of 10 children are now cured. Unfortunately, this has not been the case for all patient groups. The subset of ALL that occurs in infants (babies aged one year or younger at the time of diagnosis) continues to carry a dismal prognosis. What makes this group of patients so hard to treat has baffled doctors and leukaemia biologists alike for many years.
A particularly striking example is that of Mixed Lineage Leukaemia (MLL) gene-rearranged ALL. This results from a chromosomal rearrangement, most commonly fusing the MLL gene on chromosome 11 with the AF4 gene on chromosome 4 to create a new cancer-driving protein, MLL-AF4. What sets MLL-rearranged ALL apart is that, unlike most other cancers that require additional mutations in multiple genes, this single driver mutation is sufficient to cause a leukaemia. And yet, even within MLL-rearranged ALL, where patients have the same driver mutation, infants have a uniquely aggressive form of the disease, with significantly worse survival outcomes compared to older children. We therefore asked ourselves what factors could cooperate with the MLL-AF4 protein to cause a more aggressive disease in infants and, perhaps more importantly, should we be treating MLL-rearranged infant-ALL as a distinct subset of ALL?
Leading up to this project, very little was known about how age-related differences in the transcriptomic profile of MLL-rearranged ALL related to the developmental stage of the cells the leukaemia arises in. We therefore compared gene expression data in MLL-AF4 infant-ALL patients (≤1 year old) to MLL-AF4 childhood-ALL patients (>1 year old), which revealed a distinct transcriptional profile in the infant-ALL patients. One thing that is well known about MLL-rearranged infant-ALL is that the disease arises before birth. Human fetal haematopoietic stem and progenitor cells (HSPCs) display distinct gene expression programs, which are shut down after birth; and when we compared these fetal-specific genes to our MLL-AF4 infant-ALL-specific genes, we found that a surprisingly large proportion of the infant-ALL-specific transcriptional profile can be explained by maintained expression of fetal genes that should have been shut down. This suggested to us that the cell type in which the leukaemia first arises plays a major role in determining the biology of the leukaemia, which is likely to then affect patient outcomes.
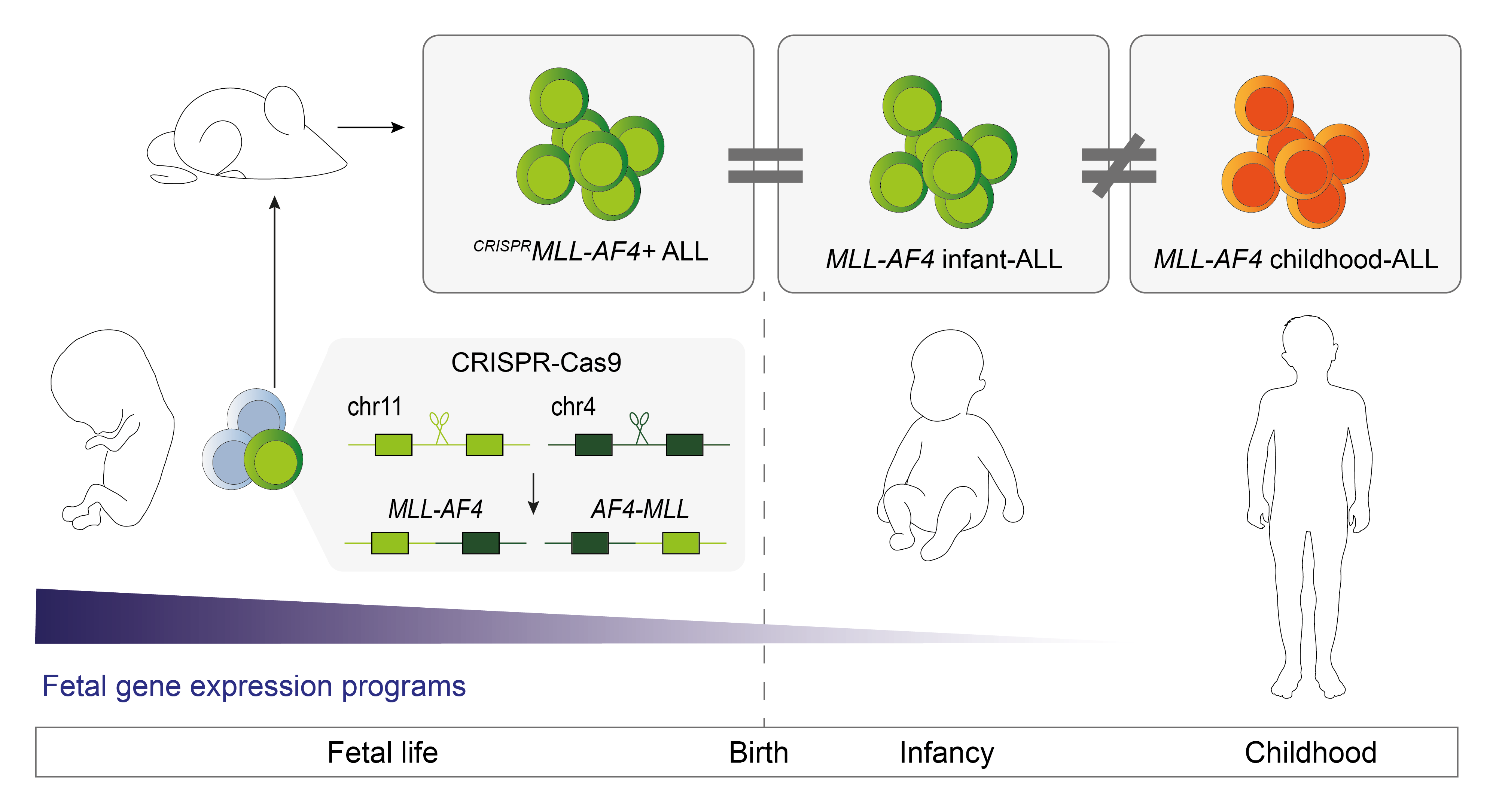
As a field, we have struggled to gain a deeper understanding of MLL-rearranged infant-ALL due to a lack of faithful models of the disease. Despite a 20-year history of studies aiming to model MLL-AF4 ALL, none has accurately captured the unique molecular and phenotypic features of infant-ALL. We hypothesised that it would be essential to express the MLL-AF4 oncoprotein in human fetal HSPCs in order to faithfully model infant-ALL. Therefore, we used CRISPR-Cas9 genome editing in human fetal HSPCs to induce the MLL-AF4 chromosomal rearrangement, then transplanted these edited cells into mice. Strikingly, the edited cells gave rise to an aggressive ALL, where cancer cells infiltrated the brain and other peripheral organs, showed resistance to chemotherapy and expressed fetal-specific genes; exactly what we see in MLL-rearranged infant-ALL. The molecular profile of the ALL we generated was also very distinct from MLL-rearranged childhood ALL, further confirming that we had truly recapitulated infant MLL-AF4 ALL.
Together, our findings provide greater insight into the behaviour of the MLL-AF4 protein, which appears to be dependent on the cell context in which it finds itself. Our work also highlights the fact that MLL-rearranged infant-ALL is a molecularly and clinically distinct subset of leukaemia, even compared to MLL-rearranged childhood-ALL. Maintenance of human fetal gene expression programs is likely to contribute to the more aggressive presentation of this leukaemia in infants, and may represent novel targets for the treatment of MLL-rearranged infant-ALL, which can now be tested in our faithful, infant-ALL-specific preclinical model.
Highlight of this project: The best part of this project was the collaboration between different research groups (Halsey, Roberts, Milne, Roy labs), clinical scientists in NHS diagnostic labs, clinicians treating children with ALL, and the generosity of patients and their families in donating samples for research. We very much enjoyed the interdisciplinary interactions, which emphasised to us how we are all striving towards a common goal: to improve the lives of children with leukaemia.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in