Silent influencer: How non-antibiotics tip the microbial balance of the gut microbiome
Published in Ecology & Evolution, Microbiology, and Biomedical Research
It would only be a slight exaggeration to say that everyone and their mother has heard about the microbiome. Our friends and family often ask us about how the microbiome is linked to their health. One of the analogies we often use is that of a forest, a garden or any other ecosystem; the gut microbiome is, after all, just another ecosystem. The gut microbiome provides us with many services—direct or indirect benefits that result from the dynamics of the species present. They include the fermentation of dietary fibers, the priming of the immune system and, important for this story, the resistance against invasion by gastrointestinal pathogens, such as Salmonella.
We are all aware of the detrimental effect some foreign substances can have on an ecosystem. In the case of a forest, it can be a pollutant or a pesticide; in the case of the gut microbiome, some of the medications we consume can have negative consequences. Yes, the negative side-effects of antibiotics on the microbiome have been known for years, but only recently the scientific community began to understand how human-targeted drugs can also lead to the disruption to the microscopic members of this ecosystem.
Given these observations, the logical next step was to ask: do human-targeted drugs disrupt the ability of the microbiome to resist the proliferation of invading pathogenic bacteria? This was the basis of Lisa Maier’s independent junior research group at the University of Tübingen, the backbone of her first successful research grant, and marked the starting point for the project taken on by the first team members.
From idea to grant to paper: growing the lab and the project together
From the start, collaboration defined our working style. Every Monday we gathered around the whiteboard, brainstorming together. All ideas were valued equally, whether it came from a technician, PhD student or postdoc. Lisa taught us how to work under anaerobic conditions, and we performed the first experiments together to reach a standard we were all happy with. We faced many challenges, but we tackled them as a team—troubleshooting in front of our whiteboard. Despite being a small group, we managed to develop a synthetic bacterial community mimicking the actual gut microbiota. Together we set up high-throughput screenings and handled new challenges when we began working with mice.
The lab grew, and the project grew along with it. As time went on, Lisa secured additional funding, the lab expanded into a full professorship, and new researchers joined the team. This brought new capabilities, such as a much-needed second anaerobic chamber, and fresh input from new lab members. Streamlining all bacteria, medications, and experiments turned out to be trickier than we had imagined. Then came creating clear, easy-to-understand figures from complex data. Fortunately, we managed to sail together through the ocean of data we had obtained.
Four years later, we were finally ready to submit our manuscript for the first time. We knew that our findings would shed light on the previously overlooked side effects. But a story is never truly finished, and the insightful comments from our reviewers helped us improve it. While the long revision process gave us a few gray hairs, it also brought unexpected new insights and collaborations, which helped us refine our story. You should have seen the fun we had imaging our synthetic gut microbial community with a scanning electron microscope for our cover artwork submission!
The pillars of colonization resistance
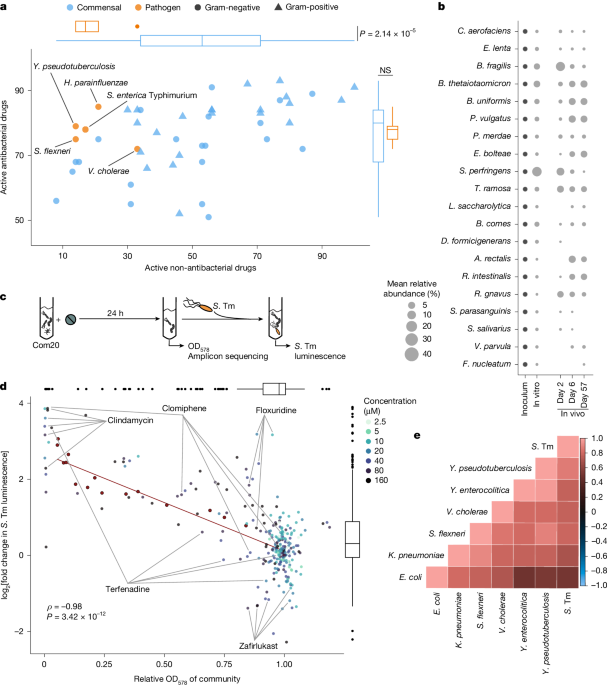
From this sometimes challenging but always engaging project, we learned that about one third of the drugs we tested lead to an increased growth of Salmonella and other pathogens in vitro. A test tube is not a gut, so we moved to mice experiments to test whether we observed similar things in a system that includes the actual body of a host or a more complex microbial community; these results were largely consistent with the in vitro data.
Now, you might ask 'how are human-targeted drugs affecting commensal bacteria?'. The answer, like almost everything in life, is not simple. In our gut ecosystem there is an enormous number of microbial species, very different from one another. These species can have very different interactions with the immense chemical diversity of medicinal drugs, therefore, we cannot point to a common way by which all drugs alter the microbiome. However, our results suggest that most drug-microbe interactions converge into three general groups: they can inhibit the growth of gut bacteria, leading to a loss of microbial biomass; they can alter the composition of the microbiome—who is there and how many of them; or they can target specific microbes that compete directly with the pathogens for nutrients. One, or a combination of these factors, can poke holes in the microbial firewall in the gut, allowing pathogens to colonize.
So, what now?
Knowing what we know about the damaging effect certain medications can have on the microbiome, what would we say to our friends and colleagues? First, don’t panic. Even though there can be microbial side effects, these are generally outweighed by the benefits of the appropriate treatment in clinical practice. The advice they receive from medical and nutrition professionals is key for making informed dietary and health choices. We are only starting to understand the underlying dynamics that shape the response of the microbiome to drug exposure. Therefore, our advice goes to medical and pharmaceutical professionals: the microbial side of things should also be considered when designing, evaluating, and using drugs and formulations, so that unwanted interactions are minimized. We expect this gut-neutral approach to gain traction in the future. In the meantime, we’ll continue to study how drugs interact with our gut ecosystem.
Jacobo & Anne.
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in