Simulation-Experiment loops to understand yeast colony growth
Published in Microbiology and Protocols & Methods

Microbes grow all around us forming biofilms and sometimes funny looking colonies on various food sources, even in our fridge. The key concepts on how microbial communities grow were laid down in the '70s and since then we have measured colonies of many kinds of microorganisms in laboratories for various tests. Still, we do not have a thorough quantitative characterisation of the factors controlling colony growth. This is getting even more relevant when we want to consider multiple organisms in a colony and understand mixed community behavior.
It all started at the local store buying bakers’ yeast, when eager (and a little impatient) students could not wait more for the final authorization of our shiny newly established laboratory and decided to start testing the tools with the safe, commercial variant. With that, the Yeast Systems Biology Research group was established merging computational modeling and experimental biology profiles of our faculty. This was not the last unforeseen challenge along the way where little creativity and collaboration with more experienced members of other labs came handy. We are grateful to all the support from Irene Stefanini, Duccio Cavalieri, Marti Aldea, Andrea Ciliberto, Gábor Balázsi, Peter Thorpe, and many others, who helped us to tackle challenges while acquiring basic and more advanced technical skills.
We originally started to work on a model describing interactions between yeast strains, but soon we had to realize that we could not build a model on earlier established growth measurement protocols, as we could not find one that takes into account all characteristics of yeast colony growth. Thus we have decided that before going to investigate interactions between multiple strains, we establish a modeling + experiment framework, which enables us to quantitatively investigate colony growth.
We tested thoroughly how changes in growth conditions affect cell proliferation and colony formation. We combined experiments with mathematical modeling to reveal how far a model, based on our current knowledge can capture the observed growth patterns. Our results revealed that colony growth is controlled by the initial spread of microbes, the wetness of media and diffusion of nutrients. The presented model can be used to simulate the growth of colonies at various initial settings and reveals which cells proliferate in a growing colony. Our results can help researchers to design and control colony growth experiments much more precisely than before.
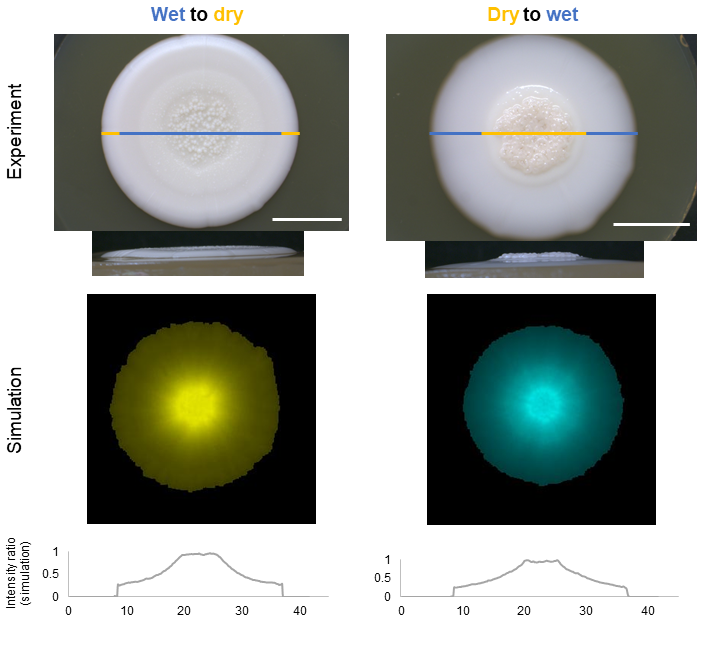
Figure 1 Example of colony growth in changing environments Colonies were grown in alternating wet and dry conditions by changing agar wetness under the colony after one week. Top and side views are shown for each scenario. Pictures were taken with a stereomicroscope. The agent-based simulations and their intensity ratios (based on the number of cells in the colony) of agent-based simulations in the cross section are shown below after the simulated 2 weeks long growth period. Scale bar represents 10 mm.
By creating this tool, we have started a long journey. There are many applications and ideas, which force us to further develop our computational tool and experimental parameterization pipeline. Besides the originally planned yeast interaction research project, we also recognized a few other applications worth exploiting. Many sources of errors, like pipette splashing when inoculating, contamination, human error or even systematic planning errors can result in inhomogeneities of the experiment and difficulties in output evaluation. In the case of a high-throughput experiment, the repetition would be costly and timely. One example for this is our 24 droplet experiment where an enlarged edge effect was observed due to the asymmetry of the samples on the sides (see Fig. 3 in the article1).
Another exciting biological question is the adaptation after environmental changes. This is an often neglected aspect, as most laboratory experiments are carried out in standardized and fixed environments. Growth differences in various environments and adaptation to perturbations can also be observed and understood in more detail with the methods introduced in our paper (wet-dry experiment Fig. 1; nutrient gradient experiment(see Fig. 5 in the article1)).
Co-authored by: Tünde Gaizer and Attila Csikász-Nagy (with input from all co-authors)
1. Gaizer, T. et al. Integrative analysis of yeast colony growth. Commun. Biol. 2024 71 7, 1–11 (2024).
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in