Single-molecule fluorescence imaging and sequencing of single synthetic polymers
Published in Chemistry
With an annual production of over 400 million tones, synthetic polymers are almost everywhere in our modern world. Synthetic polymers are generally heterogeneous, and many of them are copolymers. Their heterogeneity and sequence of monomers in copolymers are critical to polymers’ properties. In addition, structure-property relationship of (co)polymers are only defined at the bulk, average level. It is thus required to develop methods that can resolve and identify individual polymers. Here we reported optical imaging of single synthetic polymer growth at single-monomer resolution and sequencing of single synthetic copolymer chains for the first time with single-molecule fluorescence imaging.
We invented and reported a general approach, CREATS (a Coupled Reaction Toward Super-resolution-imaging) to render polymerization reactions fluorogenic and further to image it at the single-molecule level. Figure 1a shows design of CREATS for single-molecule imaging of surface-grafted polymerization, which is non-fluorogenic (Upper left). The other reaction, photo-uncaging, is fluorogenic (Lower left). By integrating the two reactions through lining the monomer and a pro-fluorescent probe, we have a coupled reaction, in which the fluorogenic signals reports the polymerization reaction (Right). We synthesized norbornene-based monomer A, carrying a BODIPY-based pro-fluorescent sidechain (Figure 1b). The uncaged product of A emits green upon UV irradiation cleavage of 2,6-dinitrobenzyl caging group. In order to image and sequence copolymers, another two monomers B and C were also designed and obtained, uncaged products of which show yellow and red emissions. Monomers with different emission colors could be differentiable and were used for imaging copolymerization.
Figure 1c presents imaging sequence for polymerization imaging using CREATS. We firstly dispersed the catalysts attached particles on the surface of microfluidic reactor. During polymerization in the reactor, monomers are supplied continuously to achieve steady-state kinetics. A UV laser uncages the monomers that are inserted into surface-grafted polymers. A delay to allow uncaged free monomers to diffuse out of the observation volume. Then a second laser is used to image the inserted, immobilized monomers and subsequently bleach them. The uncaging-imaging-bleaching cycle then repeats. All lasers are in total-internal-reflection geometry, allowing for small observation volume and better background suppression. The uncaging-imaging-bleaching cycling rate is performed much faster than the polymerization rate, so that each monomer during real-time polymerization imaging is captured. 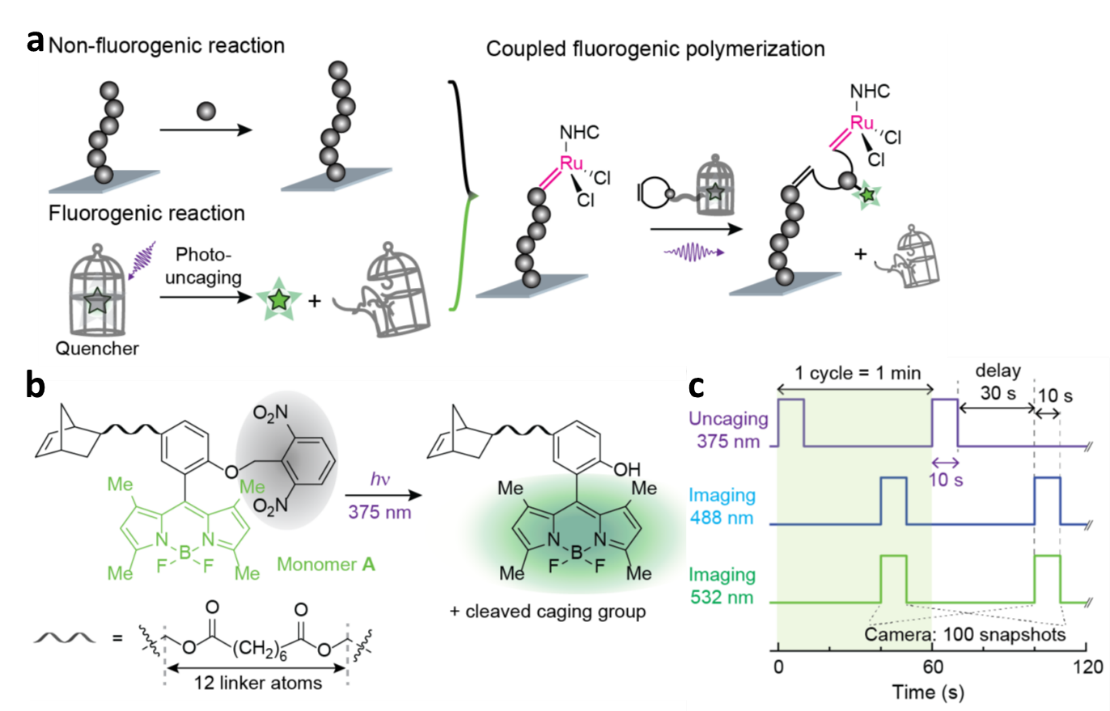
Figure 1. (a) Design of CREATS for single-molecule super-resolution imaging of ROMP at high monomer concentrations. Illustration by coupling a surface-grafted chain-growth polymerization (non-fluorogenic) reaction (Upper left) and a fluorogenic photo–uncaging reaction (Lower left). (b) Structures of synthesized caged monomer A and its uncaging reaction scheme. The caging group is shaded in grey. (c) Laser and camera timing diagram for repeated cycles of uncaging, (dual color) imaging, and bleaching the inserted monomers during polymerization.
With CREATS, we first imaged homo-polymerization of monomer A. Upon flowing the monomer solution into the reactor cell, we performed imaging using the cycles in Figure 1c. A representative fluorescence trajectory shows on-off bursts of fluorescence during polymerization (Figure 2a). The time separation τ from one burst to the next is the microscopic monomer reaction time. The distribution of τ follows a single exponential (Figure 2b), indicating a single rate-limiting step for the underlying polymerization kinetics. We also achieved nanometer-level localization of the individual polymerized monomers with the single-monomer resolution (Figure 2c). The polymerized monomers clustered around the catalyst position on the marker particle, consistent with polymerization on the same chain by a single catalyst. We also studied kinetic dispersion of synthetic polymers at single-polymer level, which reflects the heterogeneity and molecular weight distribution of synthetic polymers. Although challenging to be studied in bulk level, with the single-molecule imaging of single polymers, chain-length dependence (Figure 2d) and temporal dynamics for surface-grafted polymerization were discovered, attributable to surface electrostatics and neighboring monomer interactions, respectively. 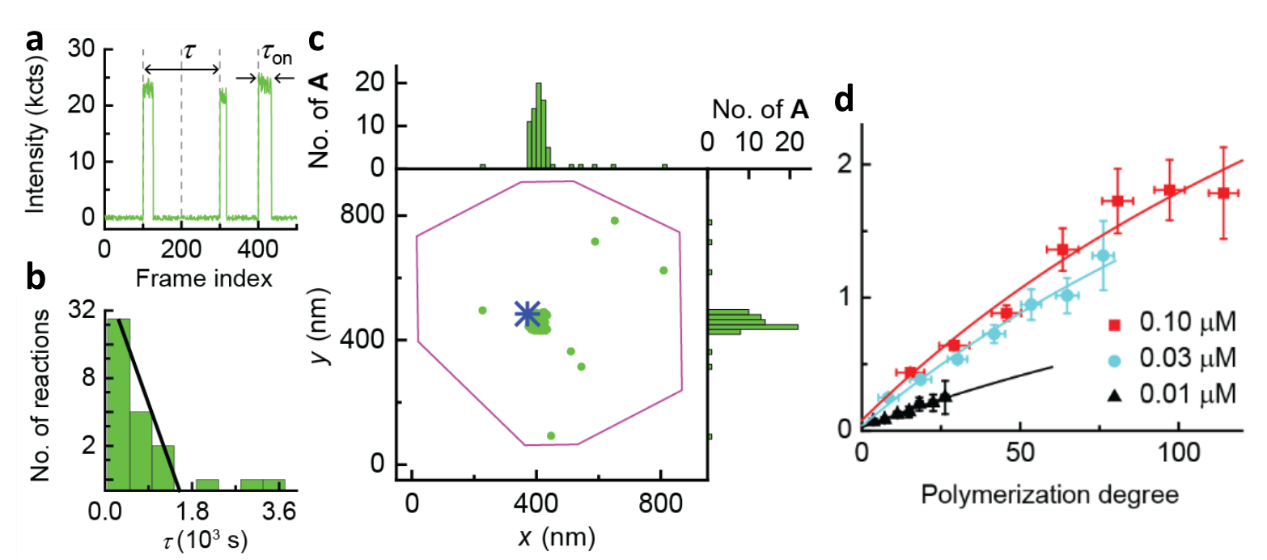
Figure 2. Super-resolution imaging of single-polymer growth at single-monomer resolution. (a) Representative fluorescence intensity trajectory during polymerization on the catalyst at [A] = 0.1 µM. Vertical dashed lines mark the start of the 488 nm imaging laser. The x-axis is plotted against image frame. (b) Histogram of τ from the catalyst. y-axis in logscale. Solid line: exponential fit. (c) Locations of inserted monomers during polymerization (one green dot: one monomer), overlaid on the catalyst location (blue star) and marker particle contour (purple line). The spatial dispersion of monomers around the catalyst is ~ 49 nm. (d) Single-catalyst TOF vs the polymerization degrees for monomer A. solid line: global fits.
We then moved to sequence single copolymers using monomers A and B as shown in Figure 3. We found that the uncaged A and B clustered around the same catalyst, supporting their copolymerization (Figure 3a). In the copolymerization fluorescence trajectory (Figure 3b), the color of inserted monomers indicates the copolymer sequence. We successfully sequenced ~ 60 synthetic copolymers individually for the first time, as no other methods existed for such tasks. The trajectories also allow us to resolve 4 different microscopic reaction time: τAA, τAB, τBB, and τBA, depending on the type of inserted monomers and following one in the growing chain. These microscopic reaction times vary but follow single exponential distributions (Fig. 3c). The reactivity ratios obtained from these times could further quantitatively predict copolymer sequence patterns, and suggest the tendency to forming block copolymers.
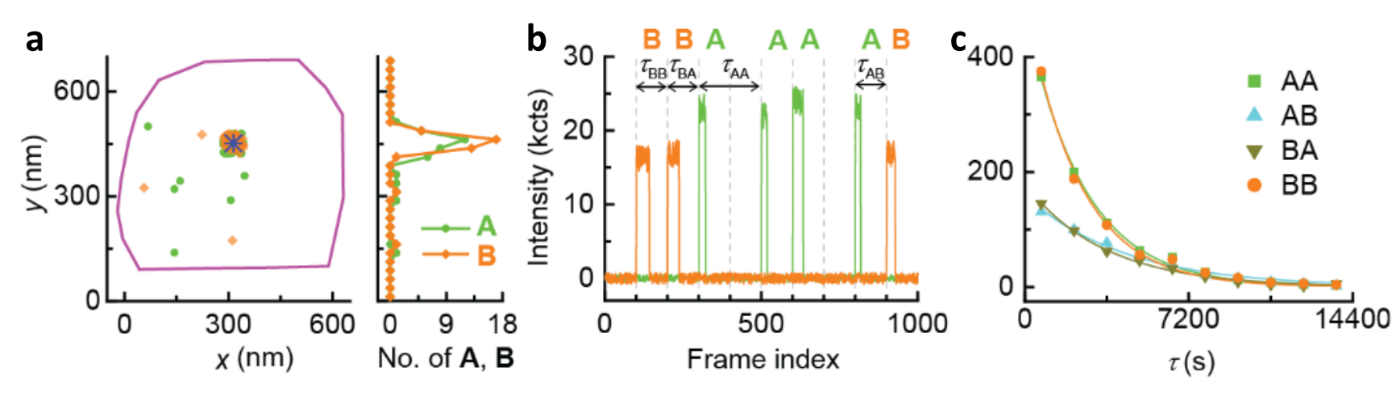
Figure 3. Single copolymer sequencing. (a) Localizations of individual polymerized monomer A (green dots) and B (yellow dots) around a single labeled G2 (blue star). Purple line: expanded contour of the marker particle from its SEM image. (b) A segment of background subtracted 2-color fluorescence intensity trajectory on the catalyst in a, showing the sequence of the grown copolymer. Vertical dashed lines: start of the 488 and 532 nm imaging lasers. (c) Distributions of 4 different microscopic monomer reaction time τ during copolymerization. Solid lines: exponential fits.
We reports the CREATS approach for sequencing of single synthetic polymers in this work, and we believe this approach is one general method as it could combat the well-known “concentration barrier” when single-molecule fluorescence method is used to image reactions. It could be extended to image any chain-growth polymerization as well as other chemical reactions, not limited to polymerization, e.g., cross-coupling reactions, if reactants are appropriately designed. In order to utilize CREATS to image reactions, there are several requirements for the reactants (monomers for polymerization in this work). Firstly, the reactants should be active for the reaction, in other words, functional groups that interfere the reaction are absent. In addition, a fluorescent probe with caged fluorescence should be linked into the reactant. We expect to develop fluorescent probes that could be quenched more efficiently by designed caged groups, which help further resolve the concentration barrier; on the other hand, the fluorescence has to be uncaged simply (in this work by a UV laser), which reports the reaction. Furthermore, the bright and photostable fluorophores are expected to improve fidelity of this approach.
We expect the CREATS approach could extend the applicability of single-molecule fluorescence imaging method that were generally limited to imaging fluorogenic reactions, and endow it to further identify and resolve challenges in various fields in catalysis, materials development, and energy conversion.
For more detailed information, see our article “Optical sequencing of single synthetic polymers” in Nature Chemistry (https://www.nature.com/articles/s41557-023-01363-2). Another blog post from co-author Xianwen Mao is at https://chemistrycommunity.nature.com/posts/navigating-the-challenge-crafting-data-analysis-pipelines-for-polymer-imaging-breakthroughs.
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in