Spatiotemporal and global profiling of DNA–protein interactions enables discovery of low-affinity transcription factors
Published in Chemistry

DNA-protein interactions are essential components of biological systems. DNA interacts with various proteins to orchestrate complicated and elusive cellular dynamics.1,2 Among the DNA-interacting proteins, transcription factors (TFs) are of particular significance since varied TFs recognize different specific DNA sequences of genes to delicately regulate the transcription and gene expression. Apparently, globally profiling of the specific DNA-TF interactions is of utmost importance regarding to deciphering the DNA-TF recognition codes and facilitates the understanding of gene regulation and phenotype in human health and disease.
Current achievements and challenge
Several elegant in vitro high-throughput strategies (Fig. 1a), such as protein-binding microarrays (PBMs) and high-throughput systematic evolution of ligands by exponential enrichment (HT-SELEX), are powerfully tools to globally measure the relative binding affinities of TF-DNA interactions using in vitro DNA and protein libraries, which have revealed preferred binding motifs (a short DNA sequence that is specifically recognized by a TF of interested) for a plethora of TFs; additionally, the motifs could in turn serve to predict high-affinity interacting TFs for a specific genomic DNA sequence.3 Alternatively, DNA-affinity-pulldown-mass-spectrometry (DNA-AP-MS)-based strategy (Fig. 1a) allows to global profile the repertoire of bound proteins in an unbiased way, which has identified binding TFs for plenty of genomic DNA with varied sequences, modifications, tertiary structures, or transcriptional status.4 All these studies significantly advanced the understanding of the regulatory roles of TFs in gene expression networks.
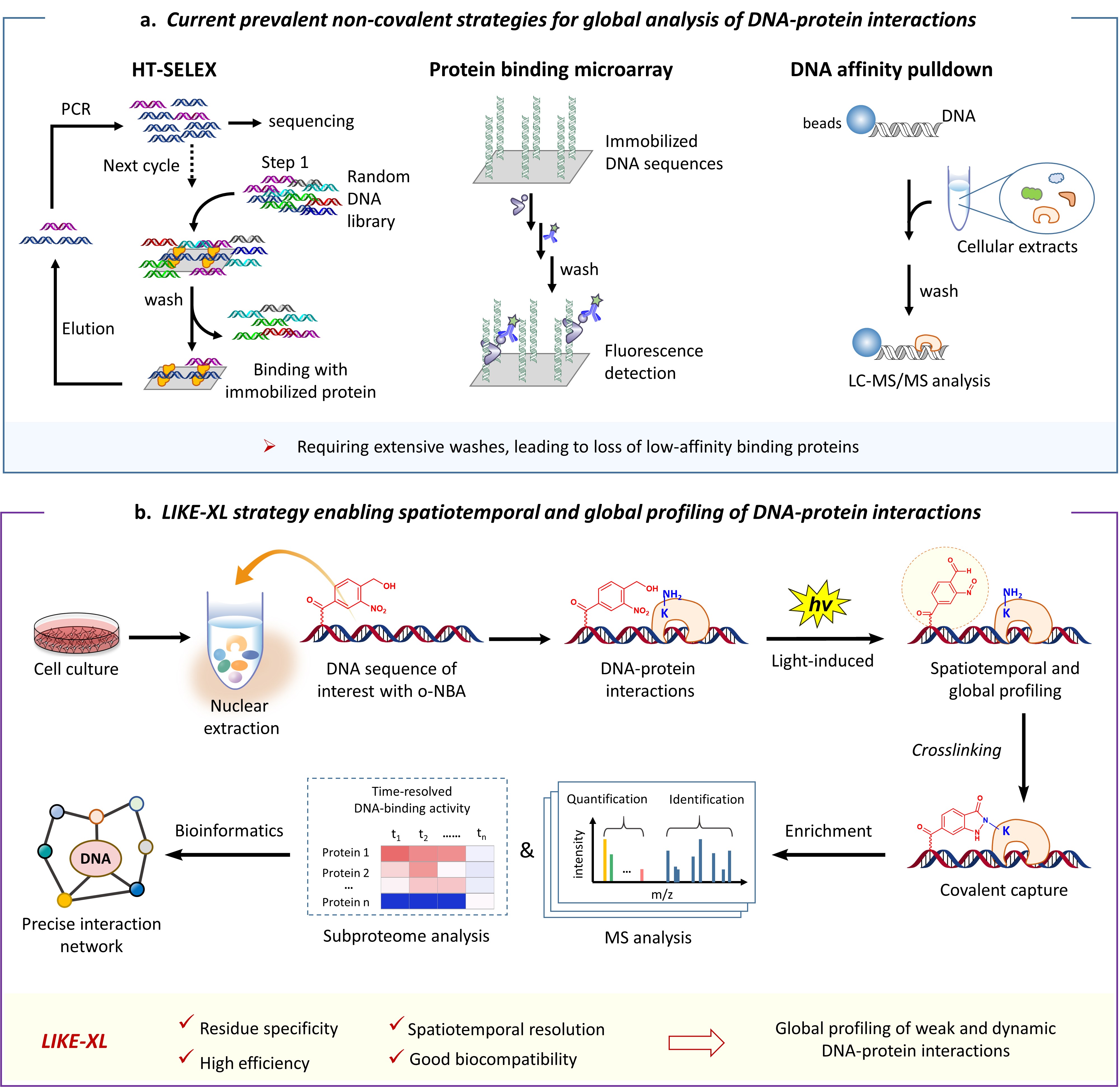
However, all of these methods are based on non-covalent DNA-protein interactions. The extensive washing and other post-process, which is required to remove non-specific binding, readily lead to loss of the weak and transient, yet specific interactions. Remarkably, mounting evidences have revealed that the weak interactions between TFs and low-affinity binding sites play critical roles in facilitating in vivo binding specificity of TFs, and thus, in regulation of gene activities and interaction networks.5 Therefore, it is a long-standing challenge and unmet need to dissect weak (or low-affinity), transient and dynamic DNA-protein interactions which occur widespread in DNA-protein complexes.
Covalent capture strategy as a potentially promising solution
Covalent capturing of DNA-protein interactions, which allows harsh post-process, offers a promising and effective strategy to identify weak and transient DNA-protein interactions. Crosslinking strategies relying on formaldehyde have long been general methods to lock interplayed DNA sequences and proteins, but the non-specific crosslinking could lead to high-level contaminants and limited spatial resolution, dramatically complicating the deconvolution of DNA-protein interactions.6 In addition, several elegant strategies have employed sequence-specific DNA baits with photo-crosslinkers or spontaneous chemical crosslinkers to capture interacting proteins.7 Nonetheless, reactive intermediates of photo-crosslinkers are non-selective to residues and readily quenched; chemical crosslinking mainly focus on labelling and bio-conjugation, or retrieving exogenous proteins with known DNA-binding activity.7 Therefore, the development of crosslinking-enabled methods to profile DNA-protein interactions (especially for the weak and transient ones) with high accuracy and efficiency, still remains challenging and highly desired in the field.
LIKE-XL strategy for covalent capture DNA-protein interactions
Lysine residues are rich in the DNA-protein (e.g. TFs) binding interfaces.8 However, methods to access lysine-selective and temporally controlled capture of DNA-protein interactions have not been explored. Very recently, we developed the light-induced primary amines and o-nitrobenzyl alcohols cyclization (PANAC) photoclick reaction for modular functionalization of diverse small molecules and native proteins,9 as well as covalently capturing protein-protein interactions in living cells,10 in which the o-nitrobenzyl alcohol (o-NBA) functionalities are converted into aryl-nitroso intermediates upon light-activation and selectively capture primary amines (e.g. side chains of lysine residues on proteins) with fast kinetics. Comparing with the DNA-protein crosslinking strategies mentioned above, the PANAC photoclick chemistry features temporally controlled manner, reliable chemoselectivity, high reactivity, good biocompatibility and long half-life under physiological conditions.9,10 Yet, the feasibility of PANAC photoclick chemistry to crosslink DNA-protein interactions is unknown and remains unexplored.
In this study, based on these insights into the PANAC photoclick chemistry, we developed the light-induced lysine (K) enabled crosslinking (LIKE-XL) strategy for spatiotemporal and global profiling of DNA-protein interactions via sequence-specific DNA baits (Fig. 1b), conferring sensitivity and accuracy. Our LIKE-XL strategy not only presents a general method for constructing oligonucleotide-dye, peptide, and protein bio-conjugates, but also shows higher effeciency than prevalently used diazirine and phenyl azide photocrosslinking methods to capture DNA-protein interactions. Moreover, harnessing unique features to capture weak and transient DNA-protein interactions, we demonstrate that LIKE-XL is capable of capturing low-affinity DNA-TF interactions and determining the binding sites previously unachieved for several TFs. Furthermore, global profiling of DNA-TF interactions via LIKE-XL presents unexpected dynamics of TF sub-proteome in response to exogenous triggers (e.g. histone deacetylase inhibitor SAHA and CDK9 protein degrader) in time-resolved manner, providing potential downstream target TFs and insights into dynamic transcriptional and protein homeostasis networks.
In conclusion, LIKE-XL strategy not only presents an effective and general method for constructing oligonucleotide-protein conjugates, but also provides a novel strategy to spatiotemporally and globally capture and identify the DNA-protein interactions with high sensitivity and accuracy. With these unique features, LIKE-XL enables discovery of low-affinity proteins via DNA sequences of interest and precise dissection of dynamic DNA binding activity profiles of proteins. In addition, the LIKE-XL offers a novel and complementary method to expand the DNA-centric profiling toolbox, to investigate DNA binding motifs previously unachieved for various proteins and to explore TFs-involved mechanism of action in drug perturbations. Moreover, as the feasibility and success demonstrated in global profiling DNA-TF interactions, the LIKE-XL is also a promising method to expand the repertoire of protein interactomes of DNA modifications. Therefore, this strategy could serve as a new tool for chemical biology, nucleic acid chemistry, proteomics, DNA and RNA epigenetics, bioconjugate chemistry.
For the full report please see: https://doi.org/10.1038/s41557-023-01196-z
References
- Hudson, W.H. & Ortlund, E.A. The structure, function and evolution of proteins that bind DNA and RNA. Rev. Mol. Cell Biol. 15, 749-760 (2014).
- Venkatesh, S. & Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Rev. Mol. Cell Biol. 16, 178-189 (2015).
- Stormo, G.D. & Zhao, Y. Determining the specificity of protein–DNA interactions. Rev. Genet. 11, 751-760 (2010)
- Wierer, M. & Mann, M. Proteomics to study DNA-bound and chromatin-associated gene regulatory complexes. Mol. Genet. 25, R106-R114 (2016).
- Kribelbauer, J.F., Rastogi, C., Bussemaker, H.J. & Mann, R.S. Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Rev. Cell Dev. Biol. 35, 357-379 (2019).
- Hoffman, E.A., Frey, B.L., Smith, L.M. & Auble, D.T. Formaldehyde crosslinking: A tool for the study of chromatin complexes*. Biol. Chem. 290, 26404-26411 (2015).
- Ivancová, I., Leone, D.-L. & Hocek, M. Reactive modifications of DNA nucleobases for labelling, bioconjugations, and cross-linking. Opin. Chem. Biol. 52, 136-144 (2019).
- Yu, B., Pettitt, B.M. & Iwahara, J. Dynamics of ionic interactions at protein–nucleic acid interfaces. Chem. Res. 53, 1802-1810 (2020).
- Guo, A.-D. et al. Light-induced primary amines and o-nitrobenzyl alcohols cyclization as a versatile photoclick reaction for modular conjugation. Commun. 11, 5472 (2020).
- Hu, W. et al. Genetically encoded residue-selective photo-crosslinker to capture protein-protein interactions in living cells. Chem 5, 2955-2968 (2019).
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in