Subdomain Dynamics Enable Chemical Chain Reactions in Nonribosomal Peptide Synthetases
Published in Chemistry, Physics, and Cell & Molecular Biology

Scientific Background: Myriads of bioactive peptides are enzymatically synthesized by large nonribosomal peptide synthetases (NRPSs), including vancomycin and teixobactin antibiotics. The sequential biosynthesis is performed by modular NRPS in an assembly-line like manner. In each NRPS module, an adenylation domain (A-domain) is responsible to activate an amino acid substrate to form an acyl adenylate intermediate that is then ligated to a neighboring peptidyl carrier protein (PCP) domain via a thioester bond1,2 (Figure 1a). A fundamentally important yet unresolved question in the NRPS field has been how these complex NRPS enzymes coordinate these stepwise biocatalysis sequences spatially and temporally. Using the A-PCP didomain in the gramicidin synthetase I (GrsA) as a model system3, we started to tackle this question by using single-molecule Förster resonance energy transfer (smFRET) spectroscopy that allowed us to directly observe individual conformations in GrsA under turnover conditions without ensemble masking4.
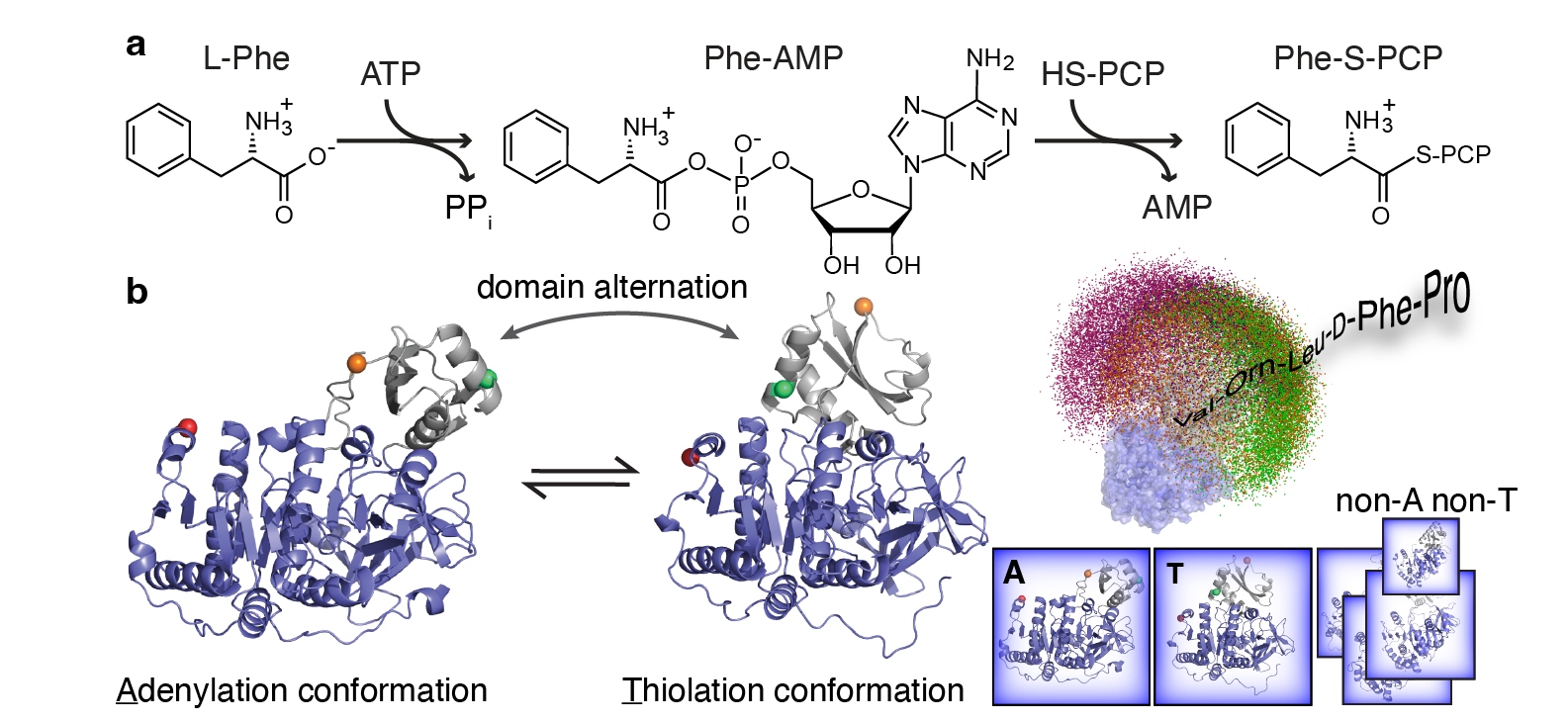
Puzzling smFRET Discovery: The very first smFRET results that I obtained as a 1st-year graduate student in the group of Prof. Haw Yang at Princeton University back in 2010 were completely unexpected: we directly observed an extended conformation that cannot be explained by two well established crystallographic A-domain structures (Figure 1b). To rule out that the labeled protein is inactive or unfolded under our assay conditions, we needed to measure the biochemical activity of GrsA mutants. Back then, standard NRPS A-domain activity assays were based on radioactive isotopes that were not accessible to the Yang group. To circumvent this issue, we developed an integrated experimental platform combining electrospray ionization mass spectrometry and high-performance liquid chromatography to measure the kinetic activity of GrsA mutants5. Working with our collaborator Prof. Dr. Henning Mootz at the University of Münster, we verified the accuracy of our kinetic assays and quickly confirmed that our smFRET mutants are indeed biochemically active. The direct observation of the extended conformation, which we later termed as X conformation, was the first milestone of this GrsA project. However, any functional relevance of the X conformation remained unsolved.
Functional Relevance of the Extended Conformations: Preliminary molecular dynamics simulations suggested that the adenylation conformation could rearrange to an ensemble of more extended conformations. Both molecular dynamics simulations and homology structural modeling also hinted that sets of new salt bridges could form in these extended conformations. We thus made two charge-reversal mutants to show that indeed perturbations of these salt bridges reduce both the population of the extended conformations as well as the biochemical activity. These results indicated the functional relevance of the new extended conformation; yet, how to place this finding in the context of the well-established adenylation and thiolation reaction sequence was still unclear.
The “Eureka!” Moment: To exchange ideas and to discuss the collective GrsA results, we had three HFSP-funded annual meetings in the U.S., Germany, and Japan from 2011 to 2013. In the last meeting hosted by our collaborator Prof. Tamiki Komatsuzaki at the Hokkaido University, we found that the intra-A-domain conformational responses to a variety of substrates and ligands from our intra-A FRET mutants are highly correlated with the bulk FRET readouts from an inter-A-PCP FRET sensor developed in Prof. Dr. Henning Mootz’s group6,7. Intrigued and enlightened by these findings, we as a group of jet-lagged tourists chatted about measuring the bulk FRET kinetics for intra-A and inter-A-PCP constructs while walking in Sapporo. Soon after the meeting, we excitedly found that both rate constants were similar within the measurement errors. These results demonstrated that intra-A-domain organizations are coupled with inter-A-PCP-didomain configurations in GrsA during catalysis. Realizing the bulk FRET kinetics manifested as a conformational relaxation process of two reversible steps, we then performed kinetics modeling to show that the X conformation is in fact an intermediary conformation between the adenylation and thiolation conformations (Figure 2a). From my perspective, this is the most challenging yet most rewarding breakthrough in this GrsA collaboration.
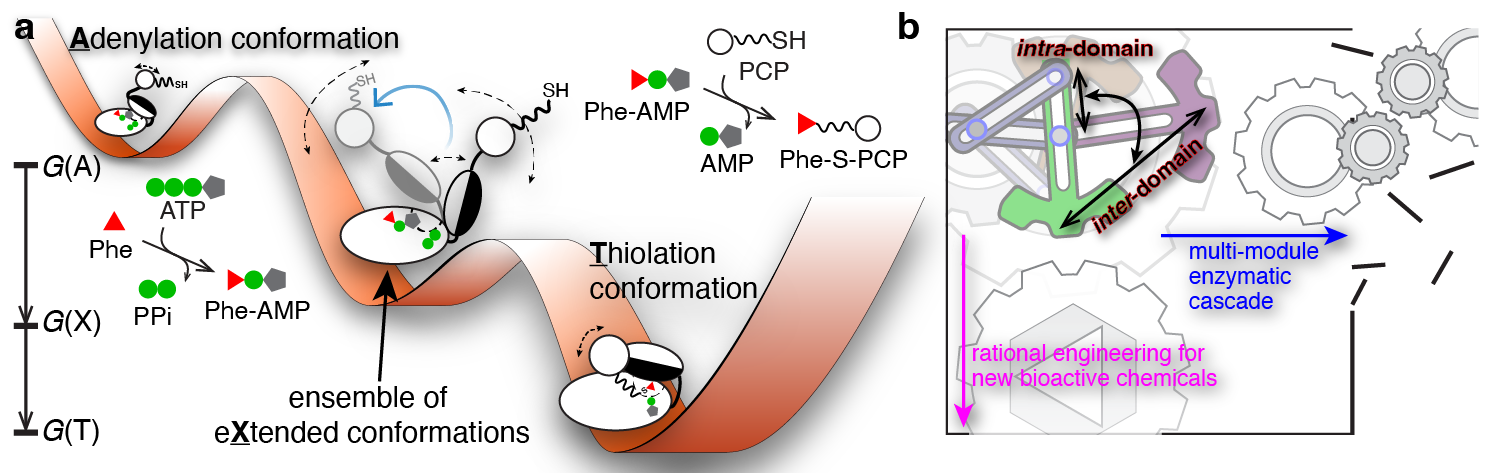
Dynamical Conformation Amplification: The last missing puzzle was how the intra-A-domain and inter-A-PCP-domain structural rearrangements are coupled. To address this challenge, Prof. Nozomi Ando (now at Cornell University) and Prof. Jhih-Wei Chu (at National Yang Ming Chiao Tung University) joined our team in 2016 and 2020 respectively, and we collaborated on integrating small-angle X-ray scattering ensemble modeling8 using smFRET dye-dye constraints and all-atom molecular dynamics simulations with dyes attached. In the end, our team showed that the X-conformation is needed to account for the measured scattering profiles of the GrsA A-PCP constructs. Importantly, our results provide experimental evidence on how intra-A-domain changes are stochastically amplified to dynamically drive inter-A-PCP conformational reorganizations, which we term as “dynamical conformation amplification” (Figure 2b).
Conclusion and Outlook: This international collaboration has been more than a decade in the making. With this work, we have established a powerful integrative platform to enable discovery of functional protein dynamics by quantitatively combining time-dependent smFRET spectroscopy, bulk FRET kinetics, small-angle X-ray scattering and molecular dynamics simulations. This work would not have been possible without the amazing perseverance and years-long contributions from all coauthors to whom I owe a heartfelt debt of gratitude. We envision that there are boundless exciting protein dynamics awaiting to be explored.
References:
- Fischbach, M. A. & Walsh, C. T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006).
- Stachelhaus, T. & Marahiel, M. A. Modular structure of peptide synthetases revealed by dissection of the multifunctional enzyme GrsA. J. Biol. Chem. 270, 6163–6169 (1995).
- Zettler, J. & Mootz, H. D. Biochemical evidence for conformational changes in the cross-talk between adenylation and peptidyl-carrier protein domains of nonribosomal peptide synthetases. FEBS J. 277, 1159–1171 (2010).
- Hanson, J. A., Tan, Y.-W. & Yang, H. in Single Particle Tracking and Single Molecule Energy Transfer: Applications in the Bio and Nano Sciences (eds Christoph Bräuchle, Don C. Lamb, & Jens Michaelis) (Wiley-VCH, 2010).
- Sun, X., Li, H., Alfermann, J., Mootz, H. D. & Yang, H. Kinetics profiling of gramicidin S synthetase A, a member of nonribosomal peptide synthetases. Biochemistry 53, 7983–7989 (2014).
- Alfermann, J. et al. FRET monitoring of a nonribosomal peptide synthetase. Nat. Chem. Biol. 13, 1009–1015 (2017).
- Mayerthaler, F. et al. Intermediary conformations linked to the directionality of the aminoacylation pathway of nonribosomal peptide synthetases. RSC Chem. Biol. 2, 843–854 (2021).
- Bernado, P., Mylonas, E., Petoukhov, M. V., Blackledge, M. & Svergun, D. I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 (2007).
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in