Super-resolution volume imaging realized by scanning single molecule localization microscopy (scanSMLM)
Published in Protocols & Methods
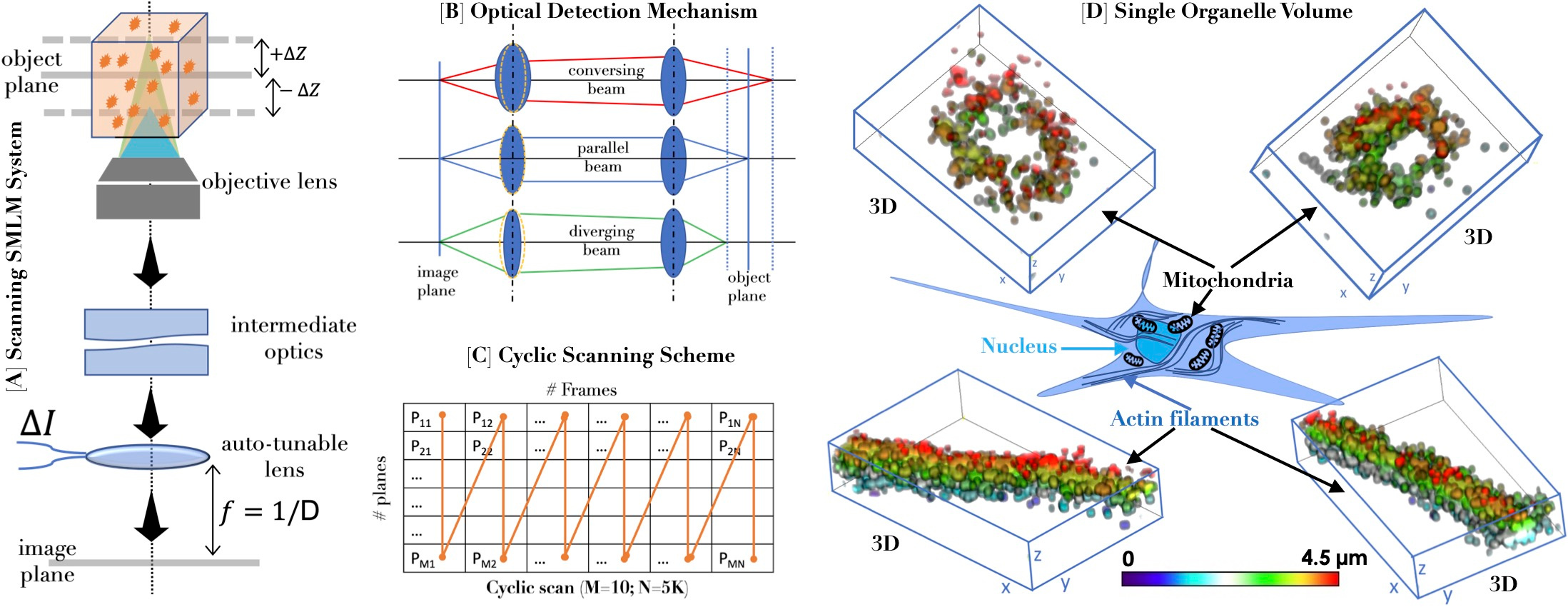
Fig. 1. [A] Schematic diagram of the scanning single molecule localization microscopy. [B] Motion-free 4f detection system. [C] Cyclic scanning scheme for bias-free volume imaging. [D] 3D distribution of single molecules on cell organelles.
Visualizing the volume of a single organelle or an entire cell with single-molecule resolution is destined to revolutionize cell and disease biology. A new single-molecule microscopy system based on scanning (termed Scanning SMLM or scanSMLM) is expected to achieve this feat [1,2]. The technique is expected to open the sky to a vast spectrum of scientists/biologist working in diverse disciplines ranging from fundamental biology to applied optical physics.
The fact that miniaturization is inevitable in modern times, especially from an application viewpoint, the future super-resolution microscopes are expected to follow suit. In this respect, scanSMLM is poised to make a mark, mainly due to its immovable detection subsystem. This is possible due to the auto-tunable 4f subsystem that does not require a bulky sample stage for axial z-scanning. The system can go down to 17.5 nm axial step (in-principle) with a speed of 200 Hz (ETL response time ~ 5ms) and image over an extensive axial range, enabling imaging large specimens without drift (for fixed stage). The system allows existing plane-by-plane acquisition (conventional scanning) or direct whole-volume data acquisition (cyclic scanning). Together with the fixed sample stage and illumination subsystem, which is static, the overall system is essentially without moving parts.
Preliminary calibration on the test sample (sub-diffraction sized fluorescent beads embedded in agarose gel-matrix) shows an impressive repeatability accuracy of > 94% for 5000 imaging cycles and the ability to scan several layers (> 10 in principle) in a specimen. It is remarkable to generate a 3D single molecule map of proteins organized on organelles (mitochondria and actin filaments) for the first time (see Fig.1). In another application related to disease biology (here, Influenza type-A), connected HA clusters are observed in a transfected 3D cell, which would not have been possible using existing SMLM techniques. This is just the tip of the iceberg, and we believe the future will see the extensive application of scanSMLM for understanding and treating other diseases, too (such as Dengue).
scanSMLM will have applications in situations that demand high-precision miniaturized design and have no scope for movable parts suited explicitly for space missions, precision measurements, and ultra-high-resolution imaging.
[Details: https://www.nature.com/articles/s42003-023-05364-2]
References :
[1] Basumatary, J., Baro, N., Joshi, P. et al. Scanning single molecule localization microscopy (scanSMLM) for super-resolution volume imaging. Commun Biol 6, 1050 (2023).
[2] Mondal, P. P. and Basumatary, J. Indian Patent Filed (#202241020010). 01 April 2022.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in