Temporal segregation of biosynthetic processes is responsible for metabolic oscillations during the budding yeast cell cycle
Published in Microbiology

Our inspiration for this study was the time-lapse-microscopy observations in single cells of the eukaryotic model organism Saccharomyces cerevisiae that the levels of cofactors, namely, NAD(P)H, ATP or flavins, oscillate during the cell cycle (see an example of NAD(P)H oscillations in the poster image above). Fascinatingly, these metabolic oscillations were found to continue in cells that stopped going through the cell cycle either spontaneously or due to different perturbations of cyclin-dependent kinase (CDK) machinery [1–3], which suggested that the metabolic oscillations are autonomous from the cell cycle. In fact, it was shown that the metabolic oscillations could exert control over the cell cycle in a system of coupled oscillators [2,4]. To understand how the metabolic oscillations exert cell-cycle control, we thought it necessary to decipher the precise nature of the metabolic flux dynamics during the cell cycle and to identify from which metabolic pathways the oscillations could actually emerge.
With the help of a novel single-cell-level dynamic perturbation method and mathematical modelling of metabolism, we provided a detailed description of various metabolic activities of yeast with respect to cell-cycle phase. Expanding our understanding of the metabolic oscillations, we discovered that a partial temporal segregation among major biosynthetic processes exists during the cell cycle and leads to changes in the fluxes of the primary metabolism (Fig. 1). Perhaps the most significant finding was that the rate of protein biosynthesis peaks twice per cell cycle, namely, in early G1 (around START) and in the middle of S/G2/M, which contradicts the current notion of either exponential or constant protein synthesis dynamics. We believe that the flux changes in primary metabolic pathways caused by the temporal segregation of biosynthetic processes would result in transient imbalances between metabolites’ production and depletion, leading to temporal changes in the metabolite levels. The effect of such imbalances would be observed in single cells, for instance, in terms of dynamically changing NAD(P)H levels. In the next paragraphs, we will present our speculations putting these findings in a broader perspective of metabolism and cell cycle.
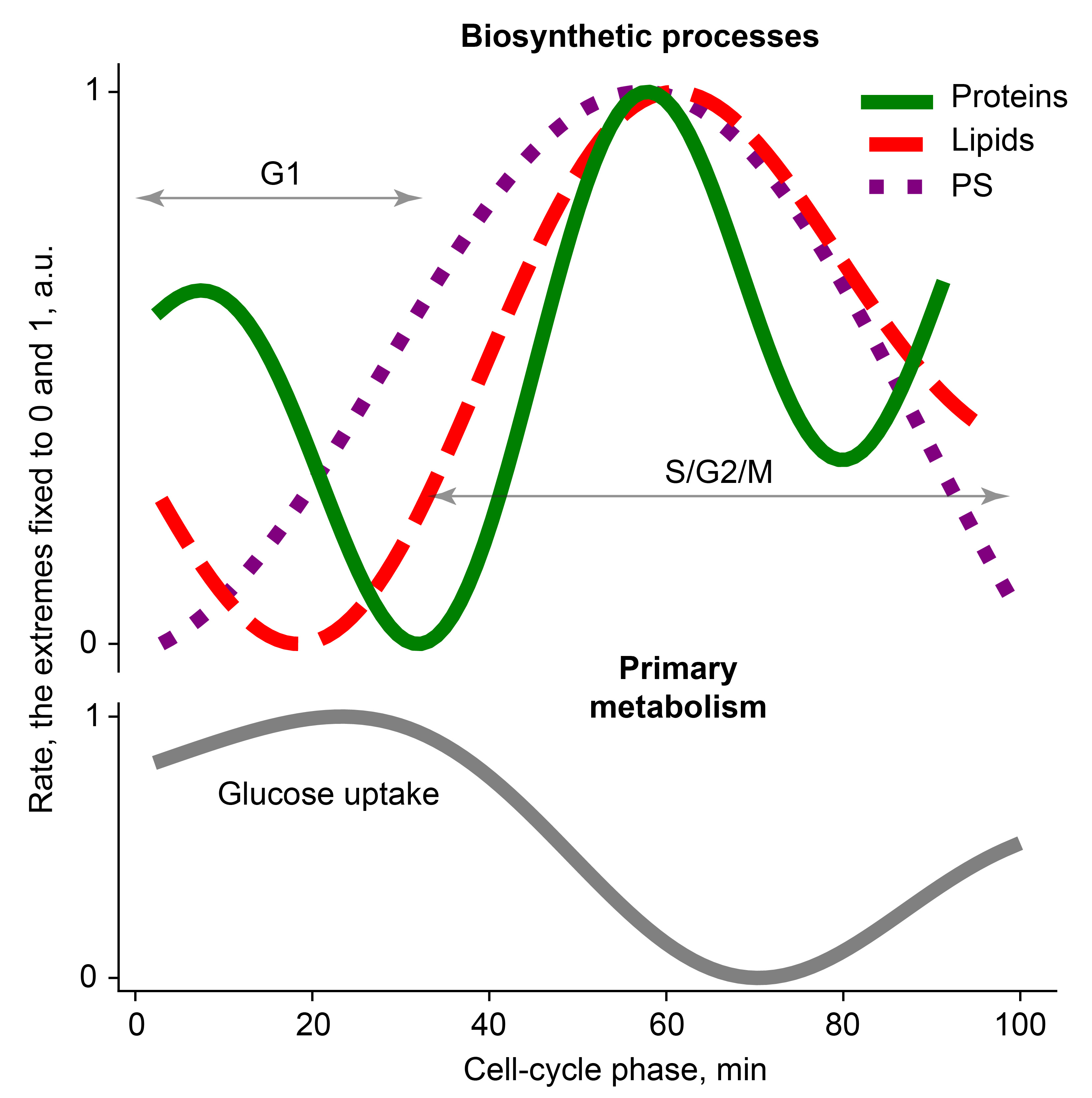
Fig. 1. The biosynthesis of proteins, lipids and polysaccharides (PS) is partially temporally segregated during the cell cycle, leading to oscillations in primary metabolism.
One of the most compelling questions is what causes the discovered temporal segregation among major biosynthetic processes. Observing that a climax of protein biosynthesis activity coincides with the slowest lipid and polysaccharide production in G1, one may conjecture that there is a negative-feedback interaction between these biosynthetic processes. Such negative feedback could be based on the competition for resources from the primary metabolism. As another idea, alternating modes of respiration and fermentation caused by the temporal segregation of biosynthetic processes (the paper’s Fig. 4b) may result in oscillations of reactive oxygen species that cause DNA damage and may thereby dynamically and differentially regulate expression of genes of biosynthetic processes, triggering their temporally segregated oscillations. DNA damage could affect gene expression, for example, by decreasing the affinity of transcription factors to their binding sites [5] or by causing RNA polymerase stalling [6].
Accumulated evidence suggests that ribosome biogenesis and production of proteins involved in translation happens primarily in G1 [4,7,8]. It was therefore rather interesting to discover that the patterns of protein synthesis and ribosome biogenesis match only partially, in G1. One could conjecture that, after ribosomes and translational machinery are made in G1, they could be partially inhibited during the cell cycle, specifically, in the S phase and at karyokinesis, when we observe decreased protein biosynthesis rate. Barkai lab has found that ~8% of the budding yeast proteome corresponds to inactive ribosomes, with this fraction being constant across various growth rates [9]. Since that study [9] was performed on the populations of cell-cycle-asynchronous cells, its conclusion could be interpreted in the way that ribosomes are inactivated during ~8% of the cell cycle corresponding to the S phase and karyokinesis.
Are there opportunities to leverage the temporal segregation of metabolism in biotechnology and medicine? If one works with a yeast cell factory and is interested in elevating the yield and production rate of, say, ethanol, perhaps it would be an idea to attempt prolonging the G1 phase in the population of cells as, according to our findings, the fluxes of glucose uptake, glycolysis and ethanol production are highest in this phase of the cell cycle (the paper’s Fig. 4b). The phenomenon that different metabolic activities are allocated to certain cell-cycle phases could potentially be considered also while tackling infectious deceases caused by unicellular pathogens. If a drug designed to disrupt a particular metabolic process essential for a pathogen is given to it during the cell-cycle phase when this process is not active, then, theoretically speaking, the pathogen could not be strongly affected and would have time to adapt before the targeted metabolic process is activated fully.
The cell-cycle machinery is likely not responsible for the temporal segregation among biosynthetic processes since, according to recent studies [1,3], metabolic oscillations manifesting in NAD(P)H, ATP and flavin dynamics persist when the cell cycle is arrested. We envision that a dynamical system of biosynthetic activities, partially segregated in time and causing the oscillations in the primary metabolism, is autonomous from the cell cycle, i.e., able to reproduce itself even beyond normal cell-cycle progression, due to the speculated negative feedback or other mechanisms. As a clue to this idea, we did not observe the NAD(P)H oscillations when the cell cycle is arrested AND either protein, lipid or polysaccharide biosynthesis is inhibited (the paper’s Fig. 5e).
We are now tempted to ask: Could it be that metabolism had already been capable of governing growth and reproduction of the cell in early evolution before an ancestor of the modern CDK machinery appeared (metabolism as a non-CDK controller [10])? Will we be able to create a synthetic cell that grows and replicates itself without a cell-cycle machinery but relies in this regard only on metabolism? Further research is needed to answer these exciting questions.
References:
- Papagiannakis A, Niebel B, Wit EC, Heinemann M: Autonomous Metabolic Oscillations Robustly Gate the Early and Late Cell Cycle. Mol Cell 2017, 65:285–295.
- Özsezen S, Papagiannakis A, Chen H, Niebel B, Milias-Argeitis A, Heinemann M: Inference of the High-Level Interaction Topology between the Metabolic and Cell-Cycle Oscillators from Single-Cell Dynamics. Cell Syst 2019, 9:354-365.e6.
- Baumgartner BL, O’Laughlin R, Jin M, Tsimring LS, Hao N, Hasty J: Flavin-based metabolic cycles are integral features of growth and division in single yeast cells. Sci Rep 2018, 8:18045.
- Litsios A, Huberts DHEW, Terpstra HM, Guerra P, Schmidt A, Buczak K, Papagiannakis A, Rovetta M, Hekelaar J, Hubmann G, et al.: Differential scaling between G1 protein production and cell size dynamics promotes commitment to the cell division cycle in budding yeast. Nat Cell Biol 2019, 21:1382–1392.
- Hahm JY, Park J, Jang E-S, Chi SW: 8-Oxoguanine: from oxidative damage to epigenetic and epitranscriptional modification. Exp Mol Med 2022, 54:1626–1642.
- Gyenis A, Chang J, Demmers JJPG, Bruens ST, Barnhoorn S, Brandt RMC, Baar MP, Raseta M, Derks KWJ, Hoeijmakers JHJ, et al.: Genome-wide RNA polymerase stalling shapes the transcriptome during aging. Nat Genet 2023, doi:10.1038/s41588-022-01279-6.
- Tu BP, Kudlicki A, Rowicka M, McKnight SL: Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science 2005, 310:1152–8.
- Blank HM, Perez R, He C, Maitra N, Metz R, Hill J, Lin Y, Johnson CD, Bankaitis VA, Kennedy BK, et al.: Translational control of lipogenic enzymes in the cell cycle of synchronous, growing yeast cells. EMBO J 2017, 36:487–502.
- Metzl-Raz E, Kafri M, Yaakov G, Soifer I, Gurvich Y, Barkai N: Principles of cellular resource allocation revealed by condition-dependent proteome profiling. Elife 2017, 6.
- Murray AW: Recycling the Cell Cycle. Cell 2004, 116:221–234.
Follow the Topic
-
Nature Metabolism

This journal publishes work from across all fields of metabolism research that significantly advances our understanding of metabolic and homeostatic processes in a cellular or broader physiological context, from fundamental cell biology to basic biomedical and translational research.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in