The DARPin platform behind ensovibep
Published in Bioengineering & Biotechnology

Explore the Research

The trispecific DARPin ensovibep inhibits diverse SARS-CoV-2 variants - Nature Biotechnology
Multiple variants of SARS-CoV-2 are inhibited by a trispecific DARPin.
When the Covid pandemic reached Switzerland in the first quarter of 2020, my colleagues Marcel Walser and Valerie Calabro had the intuition and foresight to predict that the Spike protein of SARS-CoV-2 would very quickly mutate and thus potentially outcompete both natural immunity and man-made vaccines and therapeutics.
Hence a more suitable and longer-lasting solution was desired, and upon in-depth discussion we felt that our DARPin platform could make a contribution since we had spent years making multi-specific biologics - and the trimeric spike protein of SARS-CoV-2 represented the perfect challenge for a tri-specific DARPin.
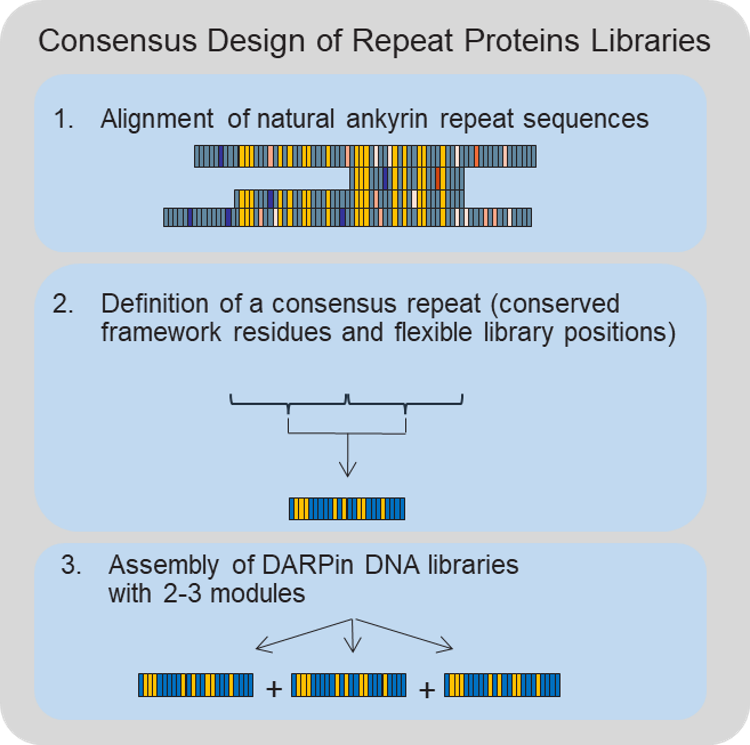
What is a DARPin? A DARPin is the designed protein equivalent of a naturally occurring ankyrin repeat protein (Binz, 2003). Nature has evolved ankyrin repeat proteins for a plethora of functions, with a common theme to use them as molecular interaction mediators, in a way like molecular glue. As such, they are also great enzyme function regulators, or inhibitors, and this is exactly why we built a company around the DARPin platform. The design and invention of the DARPin platform is described in more detail by Binz et al, 2003 (see also Figure 1). In a nutshell, we had derived the theoretical precursor of all naturally occurring ankyrin repeat proteins by simple consensus calculation (i.e. to determine the most frequently occurring natural amino acid in a given position of the protein fold). As the name states, repeat proteins are composed of the same sequence motif, stacked in tandem. To physically generate trillions of different DARPin candidates, which potentially bind to a target protein, we introduced the special feature of well-defined combinations of amino acids at the binding interface employing standard molecular biology techniques. With such a DARPin library, we then performed “molecular evolution in a test tube” experiments to obtain the best suitable lead candidates.
In late March 2020, a lead generation team was quickly called to action and agreed to work in shifts and weekends during the pandemic, and succeeded in generating very potent SARS-CoV-2 cell entry inhibitors with properties suitable for pharmaceutical drug development in a matter of just 6 weeks. The first animal data supported our belief that the tri-specific anti-SARS-CoV-2 DARPin should be moved into pre-clinical development. Consequently, larger scale manufacturing was initiated to be able to supply initial clinical studies. Since the DARPin platform relies on microbial fermentation and a platform purification process had been optimized for years, GMP material was available just 5 months after the first SARS-CoV-2 experiments had started in our laboratories at Molecular Partners AG. And the first clinical study could be initiated in Autumn 2020, a little less than 8 months after the initiation of the project.
What are the key lessons we learned in this drug development stage? First of all, we are very grateful for the absolutely pivotal collaborations with academic laboratories around the world that could be setup in a very pragmatic manner. This saved months of assay setup time and gave us access to some of the leading experts in the world in terms of viral potency assays, structural determination techniques, and animal models.
I still remember the exciting moment when the first animal model (a hamster challenged with SARS-CoV-2) read out, and we could indeed see that an antiviral DARPin could perform as well as an antibody. During this phase, we also came across a publication of a novel dwarf hamster model by the group of Jakob Trimpert at the Free University of Berlin, Germany. Within basically one e-mail and one phone call, we realized that we supported a very similar interest and embarked one of the first therapeutic explorations of this novel dwarf hamster model, with a much higher susceptibility (even reaching mortality) to SARS-CoV-2 than the traditional Golden hamster models.
Another key collaboration was initiated by a common friend, who introduced us to the group of Berend-Jan Bosch with their year-long experience in virology and especially the capability of solving cryo-EM structures. After all, “seeing” is believing. While the first experiments failed, we overcame some obstacles and realized that our DARPins bound a special conformation, similar to other antibodies described in literature. The leading investigator, Dan Hurdiss, continued to refine the calculated structures and his work allowed us to build a model of the trimeric DARPin scheduled to enter clinical development.
The most intense collaboration was built with Sylvia Rothenberger at the University Hospital of Lausanne and Olivier Engler at the Spiez Laboratory, both located in Switzerland. Profiling of countless lead candidates and later our drug development candidate against an exceedingly large number of pseudotype and pathogenic SARS-CoV-2 variants was performed in record time. This was especially important when the first Omicron variant was described in late 2021. The news came from a colleague at Novartis (our development partner) in the form of an e-mail late one evening and we had an immediate call to set the testing in motion. Thus, we could see the big advantage of our multi-specific drug candidate, ensovibep, which maintained potency on the initial Omicron variants whereas many advanced antibodies did not. Together with the collaboration with the Carol Weiss lab at the FDA and other laboratories at the National Institute of Health, Ensovibep was continuously profiled against relevant SARS-CoV-2 variants.
Overall, we learned a tremendous amount in the past two years about virology, how the international scientific community came together to quickly combat the Covid pandemic, and how collaborations can very quickly enable drug development at truly unprecedented speed. A time, I am sure, none of us will forget and we would be ready to get it going again, at any time!
Reference:
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in