The detection, biological function, and liquid biopsy application of extracellular vesicle-associated DNA
Published in Chemistry and Biomedical Research
Extracellular vesicles (EVs) are particles that are released from cells and delimited by a lipid bilayer membrane [1]. EVs can either pinch off the surface of the plasma membrane via outward budding (e.g., microvesicles with a diameter of 100–1000 nm or apoptotic bodies with a diameter of 1000–5000 nm) or can be generated inside multivesicular endosomes or multivesicular bodies (MVBs) via double invagination of the plasma membrane and then are released into the extracellular space through the exocytosis pathway (i.e., exosomes with a diameter of 50–150 nm). These EVs enclose many constituents of parent cells, including nucleic acids, proteins, and metabolites, and display a wide range of sizes. EVs are implicated in cell-to-cell communication, allowing cells to exchange components and influencing various pathophysiological processes in both parent and recipient cells [2-4].
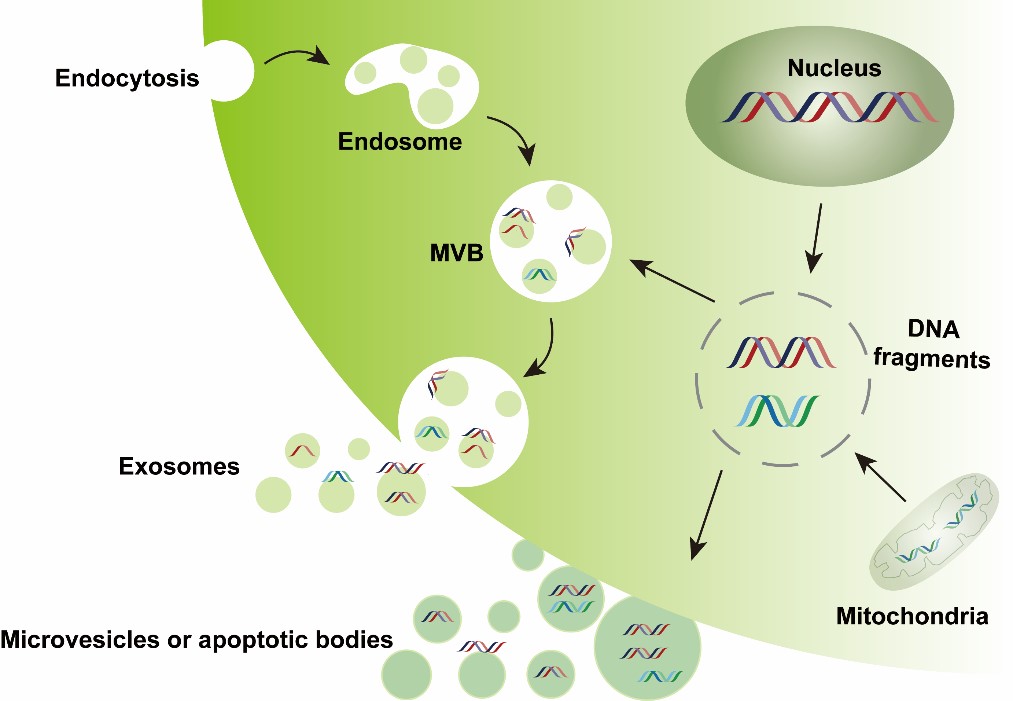
EVs are present in various tissues (e.g., the brain, melanoma, adipose, and liver) and body fluids (e.g., plasma, urine, breast milk, ascites, saliva, cerebrospinal fluid, and bile). The lipid membrane of EVs can protect their cargoes against degradation, particularly for nucleic acids. Compared with EV-associated RNA, DNA molecules were later found within EVs and have been less investigated. However, EV-DNA has attracted increasing interest, and corresponding advances are ongoing. Single-strand DNA, double-strand DNA or chromatin DNA, and mtDNA have been detected inside and/or outside EVs enriched from in vitro cultured cells and biofluids such as plasma, serum, urine, gastric juice, saliva, pleural effusion, and lymphatic drainage [5-8]. In this review, we summarize and discuss EV-DNA research advances: (1) EV-DNA detection at the population-EV and single-EV levels; (2) EV-DNA-associated biological functions, including serving as an additional gene material contributing to changes in gene expression and/or the phenotype of recipient cells and as a signal molecule activating cytoplasmic DNA-sensing pathways (e.g., cGAS-STING and the AIM2 inflammasome) in recipient cells; (3) EV-DNA-based liquid biopsy application in cancer and noncancerous diseases. This review provides potential directions or guidance for future EV-DNA investigations.
In general, EVs are first isolated from culture media or body fluids, followed by DNA enzyme digestion, EV lysis, DNA extraction, and DNA analysis. When DNA analysis is performed via nanoflow cytometry (nFCM), EV lysis and DNA extraction are not needed, but DNA staining is needed. When DNA cargoes are profiled in a single EV via hydrogel-based droplet digital multiple displacement amplification (ddMDA), EV lysis is still needed, while DNA extraction is not needed.
Heterogeneous-source DNA-containing EVs can be transferred to recipient cells and transform or affect their biological responses: (1) EV-DNA may translocate to the recipient cell nucleus and/or mitochondria and be integrated into the host genome, resulting in changes in the gene expression and/or phenotype of recipient cells; (2) EV-DNA may activate cytosolic DNA sensors of recipient cells, triggering the innate immune or inflammatory response.
EV-DNA isolated from body fluids can be used for various gene analyses (including mutation, copy number variation, epigenetic modification, etc.) with the help of diverse PCR and sequencing techniques.
Dr. Kaiyu Qian and Dr. Shan Guo are the corresponding authors of this paper.
References
1. Welsh JA, Goberdhan DCI, O'Driscoll L, Buzas EI, Blenkiron C, Bussolati B, et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J Extracell Vesicles. 2024; 13.
2. van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018; 19: 213-28.
3. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020; 367.
4. Marar C, Starich B, Wirtz D. Extracellular vesicles in immunomodulation and tumor progression. Nat Immunol. 2021; 22: 560-70.
5. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014; 24: 766-9.
6. Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A. 2017; 114: E9066-E9075.
7.Lanna A, Vaz B, D'Ambra C, Valvo S, Vuotto C, Chiurchiu V, et al. An intercellular transfer of telomeres rescues T cells from senescence and promotes long-term immunological memory. Nat Cell Biol. 2022; 24: 1461-74.
8. Zhang Y, Ding N, Li Y, Ouyang M, Fu P, Peng Y, et al. Transcription factor FOXM1 specifies chromatin DNA to extracellular vesicles. Autophagy. 2024; 20: 1054-71.
Follow the Topic
-
Biomarker Research

This is an open access, peer-reviewed journal that encompasses all aspects of biomarker investigation.
Related Collections
With Collections, you can get published faster and increase your visibility.
Oncobiome
This collection of papers delves into the burgeoning field of oncobiome research, exploring the intricate relationship between cancer and the microbiome. The oncobiome encompasses the diverse microbial communities residing in and on the human body, which influence cancer development, progression, and treatment responses. By examining these interactions, our aim is to unravel the complex mechanisms through which the microbiome impacts oncogenesis and therapeutic outcomes.
This compilation highlights cutting-edge research, offering insights into potential diagnostic markers and novel therapeutic strategies, thereby advancing our understanding of cancer biology and paving the way for innovative, microbiome-targeted cancer treatments.
This collection has been peer reviewed by the Editorial Boards of Biomarker Research
Publishing Model: Open Access
Deadline: Ongoing






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in