The Epigenetic Architects: How Polycomb Proteins Shape Zebrafish Heart and Pectoral Fin Development
Published in Genetics & Genomics
The discovery of Polycomb group (PcG) genes dates back to Drosophila melanogaster, where mutations in these genes led to striking extra sex comb phenotypes. These genes were soon recognized as master regulators of Hox genes, which orchestrate body plan specification (1,2). Over time, scientists uncovered that PcG genes are not just insect-specific; they are highly conserved across vertebrates and play critical roles in cellular differentiation, proliferation, and tissue homeostasis during early development (3,4). At the molecular level, PcG proteins work through two major complexes: Polycomb Repressive Complex 1 (PRC1) and Polycomb Repressive Complex 2 (PRC2). PRC2 initiates gene silencing by modifying histones, specifically by catalyzing the trimethylation of lysine 27 on histone H3 (H3K27me3). This epigenetic mark acts as a signal for PRC1, which further tightens the control by compacting chromatin and adding another layer of repression through histone ubiquitination. Together, these complexes ensure that genes are switched on and off at the right time, enabling proper development (5,6).
Traditional knockout models, like mice, rarely survive embryogenesis when PcG proteins are disrupted. However, zebrafish (Danio rerio) provide a unique opportunity to study PcG function in vivo. With their transparent embryos, rapid growth, and the ability to bypass early lethality through maternal mRNA contribution, zebrafish provide an unparalleled window into the role of PcG proteins during development (7). In our review, we focused particularly in heart and pectoral fin development.
The zebrafish heart is structurally simpler than that of mammals, with only two chambers—an atrium and a ventricle. Despite this, the genetic pathways guiding heart development are highly conserved. Zebrafish embryos rely on passive oxygen diffusion, allowing survival despite severe heart defects, making them a valuable model for studying cardiovascular anomalies (8). Studies have shown that both PRC1 and PRC2 play critical roles in zebrafish heart formation (9–12). When PRC1 is deficient, as seen in a mutant of PRC1 core component /rnf2ibl31/ibl31 mutants, it does not affect early cardiac marker expression but leads to severe later-stage defects, including a “heart-string” phenotype due to improper looping. This is linked to upregulated transcription factors (e.g., tbx2a, tbx3a, tbx5) and downregulation of structural genes (myl7, myh6, nppa) (9,12). Additionally, PRC1 is crucial for cardiac sarcomere assembly, maintaining heart contraction, and ensuring proper atrioventricular canal (AVC) constriction (11).
PRC2, on the other hand, is vital in the early stages of heart development. In MZezh2 mutants—where the function of PRC2’s core component, ezh2, is lost from both maternal and zygotic origins—the expression of the early cardiac marker nkx2.5 expression is greatly reduced, while another early marker, hand2, remains unaffected. This suggests that PRC2 activity for proper initiation of cardiogenesis. Mutants show impaired looping, partial expression of chamber-specific markers (vmhc, amhc, nppa), and disrupted has2 expression, leading to reduced heart size and structural defects (13). Interestingly, PRC1 and PRC2 appear to function at different stages, with PRC2 playing an early role in initiating cardiogenesis, while PRC1 ensures proper heart structure and function at later stages.
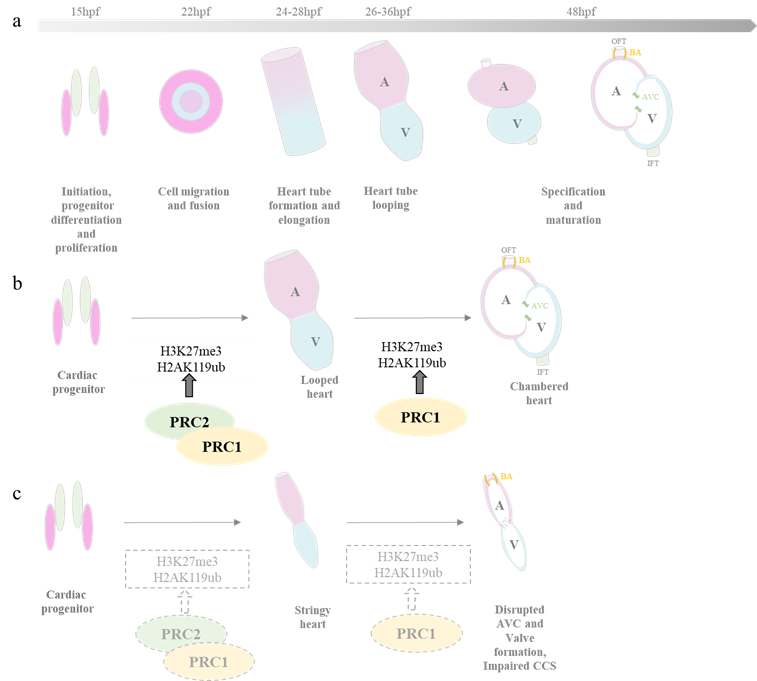
(a)Heart development in zebrafish (b) PRC2 is involved in regulating the early stages of heart development, while PRC1 contributes to the the formation and specification of the chambered heart from the looped heart structure. (c) In embryos lacking PRC1 and PRC2, the heart fails to undergo proper looping, resulting in a string-like appearance.
Beyond the heart, PcG proteins also play a vital role in zebrafish pectoral fin development. These fins share deep evolutionary similarities with tetrapod forelimbs, making them an excellent model for studying limb formation. Although zebrafish fins and tetrapod limbs differ in appearance, they rely on highly conserved genetic pathways during development (14,15). Research has shown that PcG protein deficiencies lead to complete loss of pectoral fins, underscoring their importance in limb formation. In embryos deficient in PRC1/PRC2, the initiation of fin development and the Fgf signalling pathway are impaired, leading to compromised formation of AER. Increased aldh1a2 expression leads to elevated retinoic acid (RA) signaling. Misregulation of hox genes and increased RA signaling contribute to this defect by disrupting the balance of signals necessary for fin development (9–13,16,17).
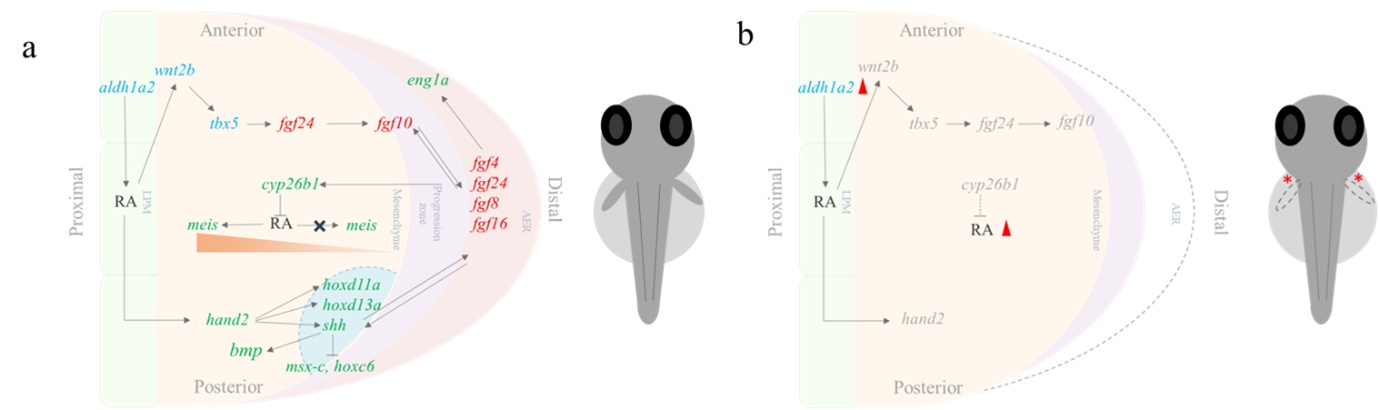
(a) Zebrafish pectoral fin development (b) Effect of PRC1/PRC2 deficiency on pectoral fin development
As deeply discussed in our review article, these findings underscore the vital roles of PcG proteins in vertebrate development. PRC2 is key for early patterning and gene regulation, while PRC1 ensures structural integrity and growth of heart and pectoral fin (18). Loss of either complex lead to severe, non-redundant developmental defects. Yet, much remains unknown. PcG mutant zebrafish fail to reach adulthood, limiting insight into their roles in maintaining organ identity and function post-embryogenesis. Future studies will be crucial to link PcG activity with broader transcriptional networks in the heart and limbs, deepening our understanding of their roles in evolution and disease.
References:
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec;276(5688):565–70.
- Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 1981 Sep;293(5827):36–41.
- Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T. The Polycomb -group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998 Nov 15;125(22):4495–506.
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006 Nov;6(11):846–56.
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science. 2002 Nov;298(5595):1039–43.
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell. 2002 Oct 18;111(2):197–208.
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993 Oct 1;119(2):447–56.
- Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 2011 Jul 15;91(2):279–88.
- Chrispijn ND, Elurbe DM, Mickoleit M, Aben M, de Bakker DEM, Andralojc KM, et al. Loss of the Polycomb group protein Rnf2 results in derepression of tbx-transcription factors and defects in embryonic and cardiac development. Sci Rep. 2019 Mar 13;9(1):4327.
- Feng G, Sun Y. The Polycomb group gene rnf2 is essential for central and enteric neural system development in zebrafish. Front Neurosci [Internet]. 2022 Sep 1 [cited 2024 Apr 20];16. Available from: https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2022.960149/full
- Peng X, Feng G, Zhang Y, Sun Y. PRC1 Stabilizes Cardiac Contraction by Regulating Cardiac Sarcomere Assembly and Cardiac Conduction System Construction. IJMS. 2021 Oct 21;22(21):11368.
- Van Der Velden YU, Wang L, Van Lohuizen M, Haramis APG. The Polycomb group protein Ring1b is essential for pectoral fin development. Development. 2012 Jun 15;139(12):2210–20.
- San B, Chrispijn ND, Wittkopp N, Van Heeringen SJ, Lagendijk AK, Aben M, et al. Normal formation of a vertebrate body plan and loss of tissue maintenance in the absence of ezh2. Sci Rep. 2016 May 5;6(1):24658.
- Sordino P, van der Hoeven F, Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995 Jun;375(6533):678–81.
- Tulenko FJ, Massey JL, Holmquist E, Kigundu G, Thomas S, Smith SME, et al. Fin-fold development in paddlefish and catshark and implications for the evolution of the autopod. Proceedings of the Royal Society B: Biological Sciences. 2017 May 24;284(1855):20162780.
- Rougeot J, Chrispijn ND, Aben M, Elurbe DM, Andralojc KM, Murphy PJ, et al. Maintenance of spatial gene expression by Polycomb-mediated repression after formation of a vertebrate body plan. Development. 2019 Sep 30;146(19):dev178590.
- Van Der Velden YU, Wang L, Querol Cano L, Haramis APG. The Polycomb Group Protein Ring1b/Rnf2 Is Specifically Required for Craniofacial Development. Riley B, editor. PLoS ONE. 2013 Sep 11;8(9):e73997.
- Kavade PS, Parab SS, Capone V, Carannante D, Ambrosino C, Altucci L, et al. Epigenetic regulation in zebrafish development: the roles of polycomb group proteins in heart and pectoral fin development. Epigenetics Communications. 2024 Dec 20;4(1):7.
Follow the Topic
-
Epigenetics Communications

Epigenetics Communications is an open-access journal devoted to the study and problematization of epigenetic principles and mechanisms in basic and translational research settings.
Related Collections
With Collections, you can get published faster and increase your visibility.
CLEPIC 2025: Advances in Epigenetics, with a Focus on Clinical Progresses, Applied Technologies and Epitarget Therapeutics
The CLEPIC 2025 Collection will showcase research presented at the Clinical Epigenetics International Conference (CLEPIC 2025), which covers the latest developments in the expansive field of epigenetics. The conference brings together researchers from the fields of molecular and clinical epigenetics, along with industry partners, ethicists, policy advisors, and other stakeholders operating in the epigenetic research and diagnostics markets.
This Collection aims to explore cutting-edge findings and innovative approaches that are shaping the future of epigenetics and its applications.
Clinical Epigenetics is devoted to the study of epigenetic principles and mechanisms as applied to human development, disease, diagnosis and treatment.
Epigenetics Communications is devoted to the study and critical analysis of epigenetic principles and mechanisms in basic research settings.
This Collection is open for submissions from all authors on the condition that the manuscript falls within both the scope of the Collection and the journal it is submitted to.
All submissions in this Collection undergo the relevant journal’s standard peer review process. Similarly, all manuscripts authored by a Guest Editor(s) will be handled by the Editor-in-Chief of the relevant journal. As an open access publication, participating journals levy an article processing fee (Clinical Epigenetics, Epigenetics Communications). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief of the journal where the article is being submitted.
Publishing Model: Open Access
Deadline: Apr 20, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in