The Frankenstein molecule: resurrecting an ancient antibiotic to create antibiotics of the future
Published in Bioengineering & Biotechnology and Microbiology

Antibiotics have saved millions of lives but, as more and more pathogenic bacteria become resistant to them, we are in desperate need of new antibiotics. Manipulating known molecules could help achieve this important goal. Since bacteria, fungi and plants represent the major source of bioactive compounds with antimicrobial activity, we could simply imitate Nature. Driven by evolution, the biomolecules we see today were optimized and refined for particular functions in a natural context. By finding out how Nature evolves complex molecules, we can learn how to manipulate and genetically engineer compounds with desired properties. This is the fundamental assumption that motivated our work.
The project idea started with Evi Stegmann and Nadine Ziemert, with expertise in natural compound biosynthesis and evolution, respectively, discussing how evolution shaped the diversity of biosynthetic gene clusters (BGCs) – genomic loci encoding the production of bioactive compounds – as we known them today. Particularly intriguing appeared the evolution of a specific class of clinically relevant antibiotics, the glycopeptides antibiotics (GPAs), because of their natural diversity. GPAs are produced by bacteria within the taxon Actinomycetes and consist of a peptidic backbone that can be further decorated with halogen, sulphate, glycosyl or methyl groups.
GPAs are synthesized by non-ribosomal peptide synthetases (NRPS), which are large enzyme complexes made up of repeating catalytic enzymes organized in sequential modules. These modules function like an assembly line so that the order of the modules specifies the sequence of the peptide backbone (Figure 1). Each module consists of catalytic domains as well as non-catalytic domains called peptidyl carrier proteins (PCPs). One catalytic domain (A-domain) selects the amino acid to incorporate at a specific position, in some cases other catalytic domains decorate the amino acid (hydroxylase (Hyd), halogenase (Hal)), and an additional catalytic domain (C-domain) incorporates it in the growing peptide chain, which is bound to the PCP. The growing peptide is then handed over to the PCP of the next module, for further amino acid incorporation and, in the case of GPAs, the crosslinking of side chains within the peptide (performed by Oxy enzymes). Finally, the crosslinked peptide backbone is released from the NRPS by the thioesterase domain (TE) and further decorated by modification enzymes, which, similar to NRPS, are encoded in BGCs.
The alternate types of GPAs found in nature contain differences in the nature of the amino acids that make up the peptide backbone, as well as in the number and/or type of modifications and crosslinks within the peptide. Despite their structural diversity, most types of GPAs share a common mechanism of action, which targets the bacterial cell wall precursor lipid II.
We asked ourselves how the ancestor of GPAs might have looked like and how GPAs may have evolved so differently. Since the diversity of GPAs mirrors the diversity of the corresponding BGCs, we first needed a phylogenetic tree to reflect the evolution of the BGCs themselves. We could not use the species tree, since the BGCs have been subject to various rounds of horizontal gene transfer within the Actinomycetes. As a consequence, the phylogeny of the BGCs does not match that of the species. Therefore, we opted for a (to date somewhat atypical) BGC centric approach to reflect the evolution of these BGCs and used a concatenated NRPS tree as a guide tree for subsequent analyses. Ancestral sequence reconstruction is a method to computationally predict from sequencing data how a gene predecessor may have looked like. Using the NRPS gene tree as a guide tree we made this prediction for all NRPS genes in the GPA BGC. Additionally, we could predict whether a gene that is seen in a GPA BGC today was already present in the ancestor BGC. This evolutionary analysis enabled us to predict the ancestral NRPS assembly line and thereby the structural core of the ancestral GPA, which we named paleomycin. We predict paleomycin would have been a complex molecule, resembling the modern GPA teicoplanin. During the course of the evolution, major rearrangements, such as gene fusion, loss or translocation occurred in the ancestral BGC that gave rise to arguably less complex, vancomycin-like GPAs.
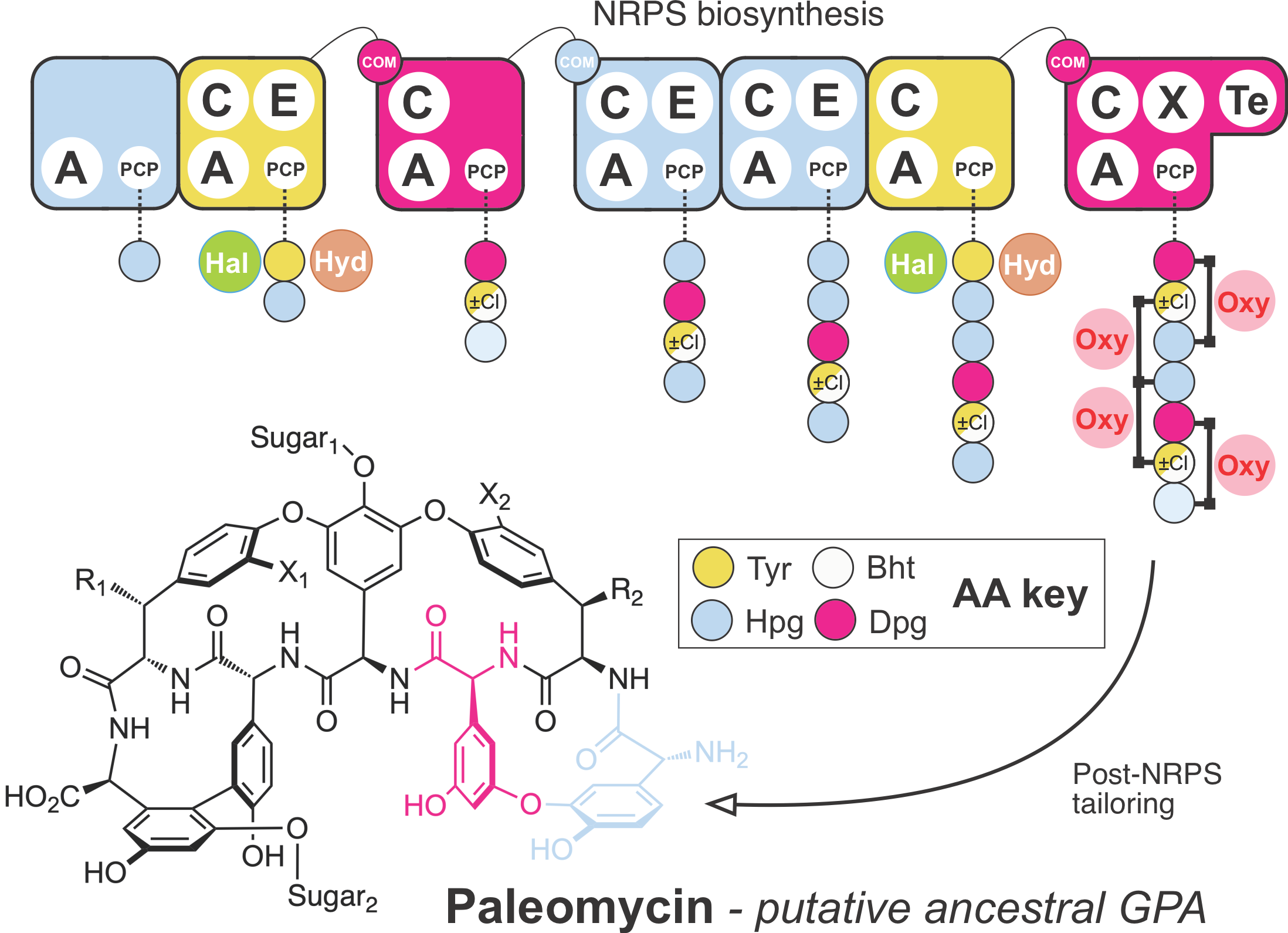
Figure 1: Overview of the modular structure of paleomycin-synthesizing NRPS. The modules of the NRPS are shown in different colors according to the amino acid (AA) they incorporated and the different catalytic domains within each module indicated as adenylation (A), condensation (C) and epimerization (E). Anchored to the peptidyl carrier protein (PCP) domain of each module is the growing peptidic chain. Green and brown circles indicate the enzymes halogenase (Hal) and hydroxylase (Hyd) catalyzing the chlorination and hydroxylation of the first amino acid, respectively. Crosslinks between amino acids are shown in the final peptide on the left, catalyzed by the oxygenases (Oxy). Com; communication domains between the individual NRPSs; X; recruitment domain for the oxygenases; Te; thioesterase, releasing the peptide from the NRPS; Tyr: Tyrosine; Hpg: hydroxyphenylglycine; Dpg: 3,5-dihydroxyphenylglycine; Bht: b-hydroxytyrosine.
When we showed the phylogenetic trees to Max Cryle, who has a longstanding expertise in biochemistry, he looked at them from a completely different perspective. Max realized that he could have leveraged the phylogenetic data to study how the NRPS domains that select the amino acids had evolved to accept different substrates and thereby produce diverse GPAs.
Encouraged in our approach by Max´s excitement for our evolutionary analysis, we were curious to see if paleomycin was really active. The only way to find out was to actually revive the long extinct compound and look for ourselves. We knew this was a high risk, high gain project. While previous studies had actually revived and expressed single proteins, no one had ever dared to completely synthesize a ~1 megadalton biosynthetic assembly line. The risk of failure was high, but at some point we decided: “Let’s just do it!” We re-performed the analysis on the DNA level for each module separately, “sewed” the modules together with intergenic sequences from Amycolatopsis japonicum MG417-CF17, the producer of the GPA ristomycin that we picked as a chassis for heterologous expression, and deleted the ristomycin NRPS genes. The process of taking things apart and putting them back to give them new life reminded us of the making of the “Frankenstein’s monster”, thus the nickname “Frankenstein molecule”. Like Victor Frankenstein, we encountered not only enthusiasm but also critical voices when we presented our work to our colleagues. While some left us really in self-doubt, we took the chance to second guess our own results and include more controls to finally be able to “show the skeptics” it’s not only hypothetically possible to produce the predicted ancestor of GPA, we did it!
Nonetheless, bringing paleomycin back to life proved to be a monstrous effort. The interdisciplinary nature of the project made the work even more difficult, as we struggled to understand each other in more than one occasion. Due to the large size and the high GC-content, among other technical problems, it eventually took us two years to get hands on a plasmid containing the synthesized sequence of the paleomycin NRPS. Troubles did not end here. The first attempts to express the ancestral GPA failed. Promoters were included/exchanged, regulators overexpressed, the “usual” attempts to enhance production of a natural compound didn’t help. The breakpoint came when we noticed how important “little” details are. Both ristomycin and paleomycin contain the non-proteogenic amino acid beta-hydroxytyrosine (Bht). However, while for ristomycin Bht is produced by a three gene subcluster on a minimal NRPS module, the ancestral assembly line first incorporates tyrosine, and then converts it to Bht by the action of a beta-hydroxylase. In initial attempts we decided to exploit the minimal NRPS naturally present in A. japonicum to incorporate Bht in the peptidic backbone of paleomycin. But only exchange of the ristomycin Bht cassette by a beta hydroxylase led to the expression of our desired Frankenstein molecule, which turned out to have antimicrobial activity against the Gram-positive bacterium Bacillus subtilis.
Around the same time on the other side of the world Max Cryle’s team was trying to understand how the NRPS changes the incorporation of the first amino acid in GPAs. Looking into the ancestral reconstruction of how these selection domains evolved pointed to a different evolutionary strategy in this case, which was the direct mutation of residues in the selection pockets of these enzymes. They then expressed the sequences for key domains that were predicted to occur during the evolution of these enzymes and could directly show how the activities of these enzymes have changed towards the formation of vancomycin-type GPAs. Next, to get insights into how these amino acid changes led to the changes in selectivity that they observed, they undertook to crystallize these domains, starting with that predicted for paleomycin all the way through to those predicted for GPAs like vancomycin. These structures showed how nature has evolved these selection pockets, which began by switching activity and then optimizing this through further mutations. Such approaches also remind us that mutagenesis approaches can still play a valuable role in the modification of peptides produced by NRPS assembly lines.
Our study shows that evolution can go the unexpected way, from more complex to more simple structures, while maintaining the same activity. We speculate this strategy might have helped GPA producers to reduce production costs by saving important energetic and molecular resources, but this really requires further investigation to understand the driving forces behind such molecular evolution.
If we have captured your interest, stay tuned: We have recently collected a dataset of over 300 GPA BGCs and plan to gain further insights into the evolution of GPAs and the construction of new GPAs by mutasynthesis experiments!
Cover art was created with BioRender by Libera Lo Presti, Leon Kokkoliadis and Johanna Stegmann.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in