The hidden lifestyle: Bacterial diversification in the biofilms
Published in Microbiology

In our group, named Bacterial Interactions and Evolution (or BIE), we are strongly interested in how bacterial populations evolve in a variety of conditions created in the laboratory. Biofilm is one of the major lifestyles of microbial communities, and studying how cells in a biofilm attach, proliferate, and evolve has become a major scientific topic in BIE. My whole PhD project involved evolution experiments conducted in different setups, such as plant roots (published previously) and nylon beads (here highlighted study).
The Bacillus cereus group, to which the bacterium we have been studying belongs to, displays diverse phenotypes. For example, some isolates show plant-promoting effects, while other isolates are defined as pathogens threathening human society. Because biofilm formation is hypothesized to be important in transmitting in their natural niches, studying how biofilms of B. cereus evolved and investigating the consequences were the best way to dissect B. cereus lifestyles.
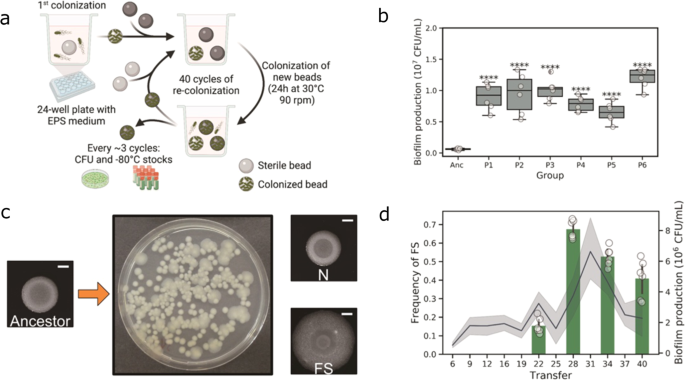
The experimental system is elegant and powerful and was initially designed for studying the biofilm evolution of Burkholderia cenocepacia during 1500 generations of selection in the lab of Vaughn Cooper. We used it as the basic method with only minor modifications. Simply speaking, it is a selection system that allows bacteria to form biofilm and later disperse to colonize a new bead. We used a similar system previously to study experimental evolution of plant roots, thus on a biotic suface.
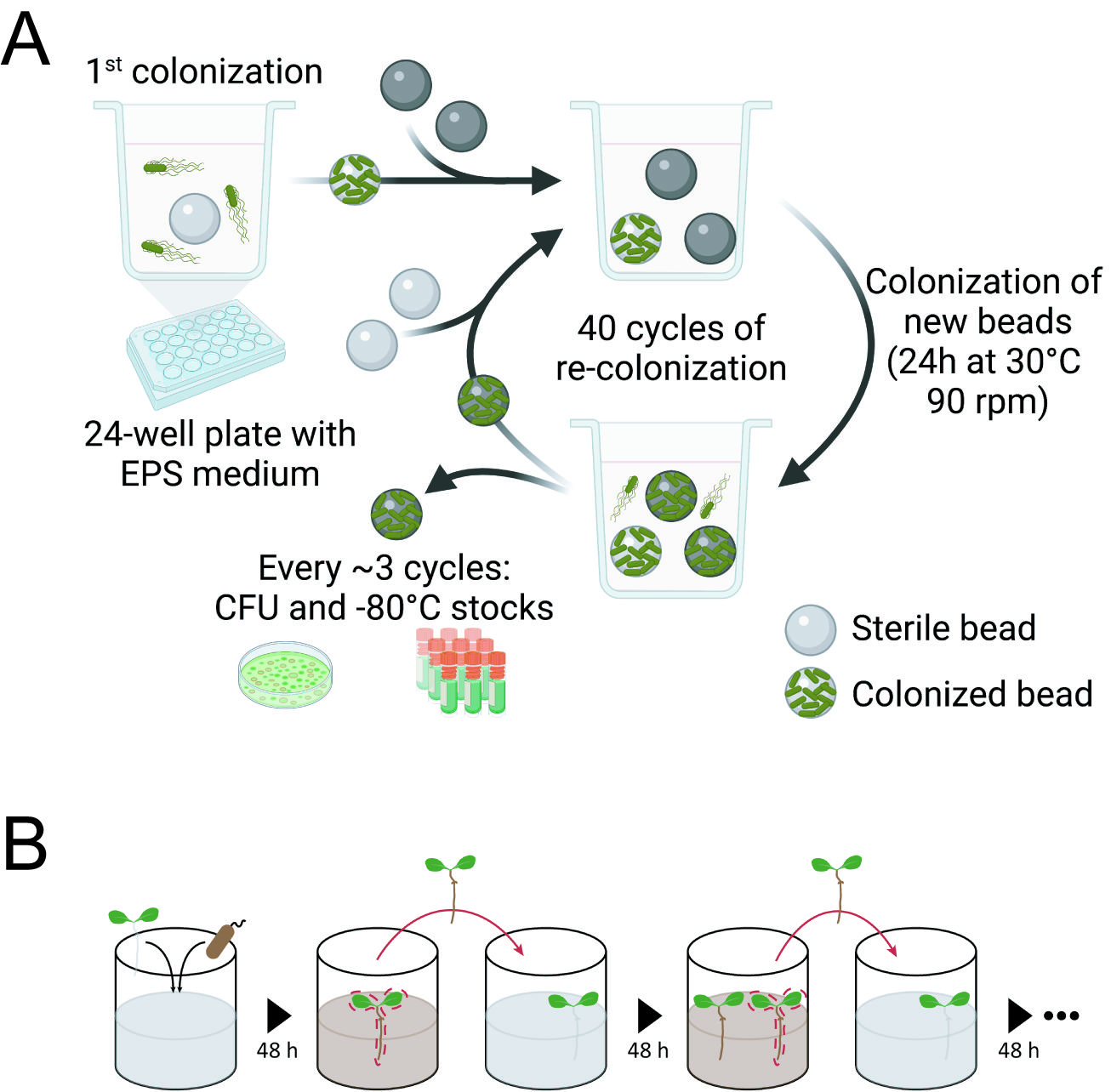
We first observed the rapid diversification of bacterial biofilms. This is a striking result regarding the fact that we did not observe morphotype diversification in the previous selection regime with plant roots. We hypothesized that this might be due to the large bacterial populations in the bead system compared with the limited cells in the plant system. The ability of B. cereus to diversify proved once again that biofilm could be an ideal model for studying bacterial community interactions.
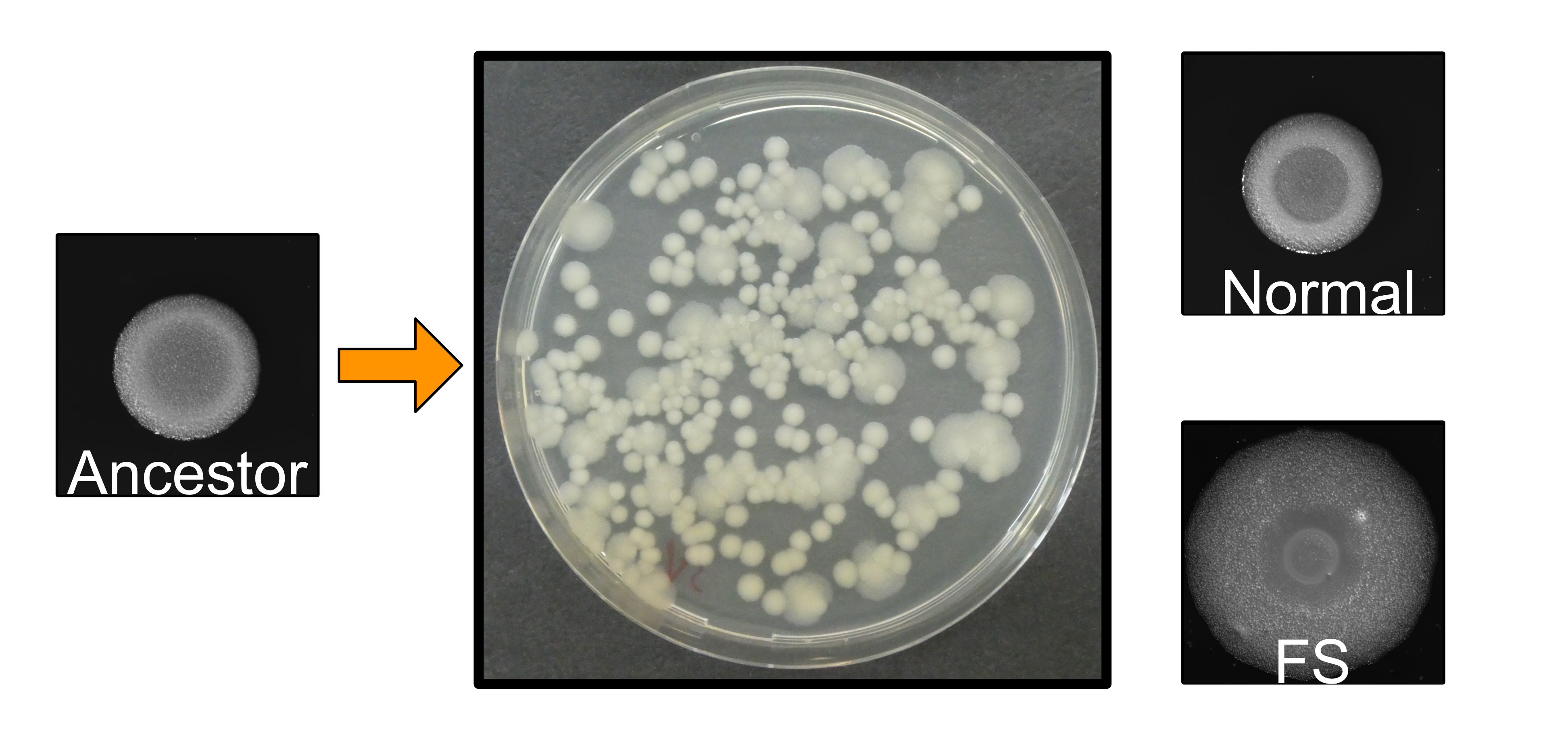
Next, we characterized the basic phenotypes of evolved isolates. The distinct one, termed "fuzzy spreader" (FS), demonstrated enhanced congo red uptake, suggesting higher production of biofilm matrix. The increased swarming and swimming ability on the tested medium indicated increased biosurfactant compound secretion and colonization ability. This was confirmed by the aggregated cell morphologies and increased biofilm formation in test tubes of FS. Moreover, we tested the growth parameters of all three isolates in the minimal medium. Intriguingly, both evolved morphotypes demonstrated less efficient planktonic growth compared with their ancestor. What is surprising is that when cultured together, these two evolved isolates exhibited a synergistic effect.
We were also interested in how evolved morphotypes would perform during a competitive fitness assay against the ancestor. Unlike the normal morphotype, the FS holds the highest fitness against the ancestor, both in colonization and re-colonization. This suggests normal isolates only adapt to the biofilm lifestyle, while the FS morphotype is a generalist that balances biofilm formation and dispersal.
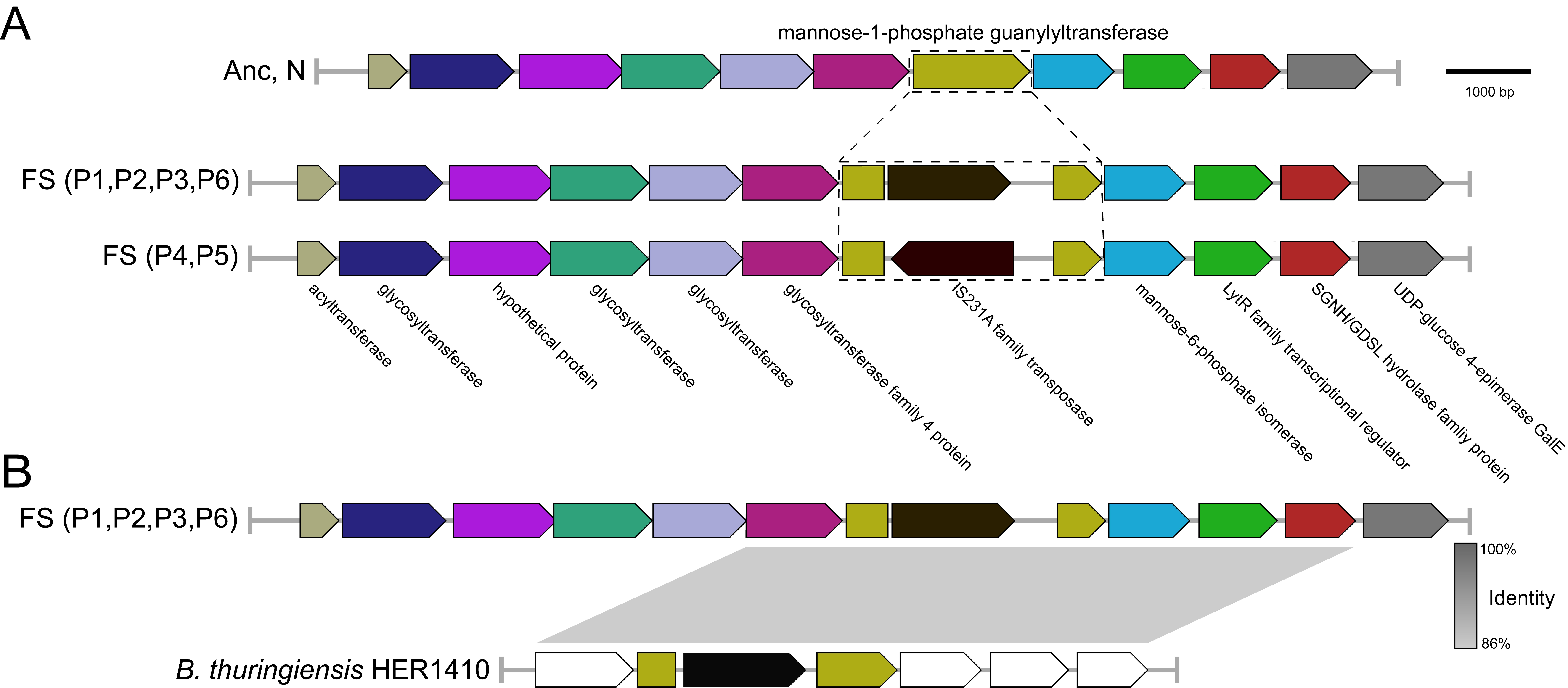
One of the most valuable products of evolution experiments is the foundation of genotype-to-phenotype correlation. In my project, we went through short-term evolution, various assays, and genome sequencing. The first touch of the Illumina data gave us confusing results and we could not clearly differenciate specific mutations connected to the FS morphotype. Thus, we turned to Nanopore sequencing, where the long sequencing reads allow us to assemble the genomes keeping also the genomic variations. To our big surprise, the gene for mannose-1-phospahte guanylytransferese was interrupted by an insertion sequence in the FS morphotype isolates, suggesting the gene is responsible for the morphotype.
Can laboratory evolution reflect what is happening in nature? To answer the question, the most interesting finding was that a similar insertion was ideintified in a homologous gene in a natural B. thuringiensis isolate. This suggests that the gene may be a genetic rearrangement hotspot related to bacterial adaptation to certain environments. It also uncovered a previously hidden role of IS elements in the biofilm lifestyle of Bacilli. The striking parallelism between the mutational outcomes of evolved and natural isolates proves that experimental evolution is a powerful tool to study bacterial genetics.
Finally, we created a deletion mutant of the guanylytransferase gene to bridge the gap between genotype and phenotype. Confirmingly, the altered phenotypes in the mutant were comparable to those observed in FS, indicating the gene was indeed responsible for the fitness and phenotypic changes.
After three years as doctoral students and a life full of assays, data analysis, and discussions, we have uncovered the first page of the evolutionary lifestyle of bacterial B. cereus biofilms. In particular, the comparative analysis of evolution studies on both plant roots and nylon beads provided valuable experimental materials for further research. The generated data paved the way to an understanding of how B. cereus biofilms might evolve in a new environment. Along the way, I’ve appreciated what the collaborators have been doing for this study. A credit goes to Xinming Xu for the RT-PCR experiments. Thanks to Gergo Maróti for DNA sequencing and analysis. Big thanks to Mikael Lenz Strube who discovered the interesting parallelism across strains, making this piece of work much more convincing and appealing to others. As always, I'd like to thank Ákos T. Kovács for initiating and steering this project in the right direction. I can always remember the consulting and communication with others for practical suggestions and ideas. Without him, this story could not be completed.
Follow the Topic
-
npj Biofilms and Microbiomes

The aim of this journal is to serve as a comprehensive platform to promote biofilms and microbiomes research across a wide spectrum of scientific disciplines.
Related Collections
With Collections, you can get published faster and increase your visibility.
Natural bioactives, Gut microbiome, and human metabolism
Publishing Model: Open Access
Deadline: May 20, 2026
Harnessing plant microbiomes to improve performance and mechanistic understanding
Publishing Model: Open Access
Deadline: Jun 01, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in