The Making of Borderlands Science
Published in Microbiology, Protocols & Methods, and Computational Sciences

Inception
In 2015, Attila Szantner (CEO and co-founder of MMOS) approached Randy Pitchford (founder and CEO of Gearbox Entertainment), with an idea to combine video games and science. Szantner and Pitchford met at the Game Developers Conference in San Francisco where Szantner presented a novel concept to integrate scientific research in the form of citizen science mini-games within major video games. The goal was to create scientific mini-games that matched the narrative, visuals, and gameplay of games with millions of players, with the hope of solving the often modest long-term engagement patterns in citizen science [1]. It was also around this time when MMOS revealed this innovative proposal [2] and began building its first collaboration with the game industry, a collaboration with EVE Online, that later culminated in Project Discovery [3].
Gearbox is known for developing successful franchises in the game industry, such as Borderlands, an action role-playing first-person looter shooter video game designed for a broad public and distributed on consoles and PCs. Over the following years, MMOS and Gearbox met several times with the primary goals of gaining traction for the project in-house, identifying potential scientific partnerships, and pinpointing the most likely video game title in the Gearbox portfolio for such a collaboration. Eventually, they agreed to deploy a citizen science activity in Gearbox’s next major title: Borderlands 3.
Early in these discussions, at a visit to Gearbox’s headquarters in Frisco, Texas, the Gearbox team was introduced to the Phylo citizen science project, led by Jérôme Waldispühl, who pioneered the gamification of genomic data analysis [4], which was publicly released in November 2010. Phylo, as a genomics scientific tool, is highly versatile in analyzing a vast range of genetic datasets, and one of its major innovations was to turn a scientific task into a casual game that can be played without extensive training: a perfect match to Gearbox games, as a scientific mini-game could be developed to fit the art style of Gearbox’s fast-paced action shooters.
The last major piece of the puzzle fell into place with the establishment of a Gearbox studio in Québec. A team of Gearbox team members from its Québec studio volunteered to work on the project and, under the leadership of Co-Studio Head Sébastien Caisse, Gameplay Director Gabriel Richard, Producer Amelie Brouillette, VP of Production Aaron Thibault and Director of Institutional Partnerships David Najjab, the project officially began.
Project setup
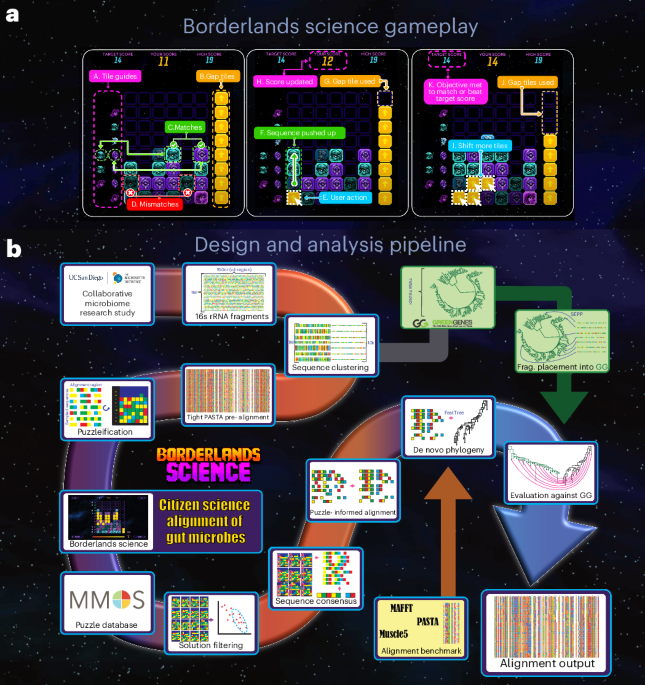
The first task was to set up a collaboration with a genomic data provider team. The field of human microbiome research emerged as a contender because of the diversity of tasks and its relevance to human health. Fortunately, the McGill team had previously engaged with The Microsetta Initiative (TMI), a human microbiome project based at UC San Diego led by Rob Knight with scientific direction from Daniel McDonald [5]. TMI is a citizen science initiative that assembled a massive public de-identified human microbiome data set with the contribution of tens of thousands of volunteers. The UCSD team embarked on the project and brought with it millions of unique and denoised 16S rRNA gene sequences that needed to be annotated.
A major challenge of such projects is the different logic and timescale of science funding, which can often become a roadblock when setting up such collaborations. That is why having established research initiatives with predictable and sustained funding, like the Phylo or the Microsetta team, was crucial in bridging this gap. As the project advanced, research agencies like Genome Canada and Genome Québec also provided crucial financial long-term support for the initiative.
In addition to securing funding, the team also had to begin developing an infrastructure for the project. Fortunately, the backend infrastructure of MMOS in the previous years had been developed to a production-ready state, thanks to an EU Horizon grant, and had proven to work well under stress during the highly successful launches of two iterations of EVE Online’s Project Discovery. This MMOS platform was designed as a highly scalable service connecting multiple research teams that have a diverse set of citizen science tasks with multiple video games by standardizing research datasets and providing an API (application programming interface) for game developers. This API does the citizen science heavy lifting, like allocating tasks to players or evaluating player contribution quality, so game developers can focus on the front-facing user experience to ensure we maximize the reach and engagement value of these science mini-games. Having a robust technological foundation for the project substantially increased the probability of a successful launch.
Building on existing achievements was a recurring theme during the project. This manifested in an iterative approach in game design where Gearbox Game Designer Gabriel Richard referenced findings from the Phylo citizen science project. But it also meant sharing knowledge with EVE Online developer, CCP Games, who by that time had years of experience introducing citizen science in a major video game and to player communities. This manifested in a three-day in-person knowledge-share session in Reykjavik among CCP, Gearbox, and MMOS experts. Such collaborations among the AAA video game industry, academic institutions, and other companies (SMEs) are unfortunately rather rare. That is why providing the game developer community with positive and fruitful examples is important.
Developing Borderlands Science
In Spring 2019, the development of the citizen science mini-game began. Immediately, the programming team implemented a mock-up of Phylo to experiment with the user experience. It quickly became obvious that the original game of Phylo was too complex and too long for Borderlands 3. This initiated a series of meetings between the game design and scientific teams, where they revisited the representation of the DNA alignment tasks and explored new gameplay options. The objectives were clear: the teams wanted a game (i) that was intuitive, (ii) with progressive levels of difficulty, and (iii) whose entry levels could be completed in less than a minute. The game designer then introduced major innovations, namely a rotation of the board that allowed them to play with the gravity of the tiles (i.e., by default the tiles are collapsing to the bottom of the gameboard, which mimics one tactic developed by participants of Phylo who often started packing all tiles on one side of the gameboard), and a decoupling of the scoring function modeling the cost of gaps as a limited resource (See Supplementary Material of the paper).
Initially, it was not clear that these changes would fulfill the scientific purpose of the underlying alignment task. Yet, the scientists saw an opportunity to revisit the information captured by the game. In subsequent meetings, the team embraced Richard’s vision of the game and embedded the scientific task into the new design. This approach forced the researchers to reevaluate the meaning of the information obtained from humans, and efficiently leverage the potential of games to build scientifically relevant experiences.
The production of a major commercial video game such as Borderlands 3 is a long process. The timeline and priorities of development were constantly evolving and required careful coordination of the departments (e.g., programming, Q&A, Art, UX). Since the development of a citizen science mini-game could not supersede the main game's development, it was critical that the scientific team worked in close collaboration with the producer and remained as flexible as possible. Unavoidably, the development of Borderlands 3 required a reconfiguring of its teams to shift priorities. Sometimes, these changes put the development of Borderlands Science on hold, often on short notice. During these pauses, regular communication between producer Amélie Brouillette, and lead scientist Jérôme Waldispühl was essential in keeping the momentum of the project and pushing forward on tasks that did not require the immediate attention of the development team. Occasional in-person visits by the researchers to the Québec studio also sustained team dynamics. Hence, the physical proximity between the academic research group based in Montréal and the game studio based in Québec City was a valuable asset for the project.
Under the hood
At the other end of the crowdsourcing pipeline, MMOS provided the infrastructure for distributing and collecting scientific data to/from the Borderlands Science puzzles in a standardized fashion. In previous MMOS projects with EVE Online, the team learned that player communities can exceed all expectations in activity, as players are relentless in the pursuit of a solution to a puzzle in a video game. Therefore, the MMOS platform was prepared to work with any size dataset circularly, providing players with a virtually limitless number of puzzles. It was important to ensure that the activity did not run out of tasks to solve. Though citizen science generally relies on the concept of distributing the same puzzle to as many different people as possible, after a certain number of players’ contributions, the quality of the community consensus did not improve. As a result, the team had to make sure to feed the platform with enough data to avoid wasting contributor effort by redirecting players to already-solved puzzles. In addition, the supervision of the data preparation phase by MMOS, which had previously deployed similar projects like Project Discovery in EVE Online, was essential, as precise figures on the amount of input data can be delicate to estimate during the initial activity peak of a game launch.
Finally, another challenge associated with the production of a project such as Borderlands Science was the need to reconcile pacing between science and industry production. A typical computational biology research project would follow its own schedule and be composed of a group of scientists and students working daily on separate tasks, meeting weekly, and addressing issues as they came. It’s also common for research projects to alternate between periods of quick progress and periods of stalling, which can last months. This pacing, however, is hardly compatible with the obligations of a game development studio, and so adapting to the production schedule was a significant challenge for the scientific team. Therefore, in the few months before launch, the team at McGill working on designing the puzzles and the analysis pipeline collaborated directly with the game producer to adopt practices from the video game industry. The team at McGill implemented precise deliverables and timelines, schemas detailing individual responsibilities to allow fast reactions to all potential launch-time problems, and assigned bug fixing and tasks in an issue-tracking software. This new way of working required substantial adaptation from the team, especially from students without prior industry experience, but this process was a strong contributor to a seamless launch. Not to mention, it also enabled these students to build connections outside academia and open new career opportunities.
In early 2020, the mini-game was ready to launch. The crowdsourcing pipeline was operational but the correctness and efficiency of the flow of information needed to be validated by the scientists. A final full-scale testing session enabled us to identify data processing features that would improve the efficiency of the computational infrastructure, such as randomizing the puzzle setup to prevent cognitive bias and avoid redundancy in MMOS databases. Since last-minute fixes are challenging to implement in large projects like Borderlands 3, it was very important for the team to have ongoing efficient communication with the game producer.
The launch
On April 7, 2020, Borderlands Science went live as planned [6]. What was not planned were the COVID pandemic and the subsequent societal disruptions. At the time, it seemed risky to push forward with the launch — a critical moment that heavily determined the success of the initiative — as the attention of the world was focused on more pressing issues. Yet, the team decided to stick to the plan, and eventually, the ongoing pandemic made the initiative even more meaningful and relevant to the Borderlands community. The launch day wrote citizen science history: that single day Borderlands Science collected five times more data than Phylo did in ten years. The Borderlands Science team believes this is just the beginning of revolutionizing citizen science in alliance with video games and gaming communities. Borderlands Science is just the start of showing how science can indeed become massively multiplayer.
Want to learn more about Borderlands Science? Check out the recent findings published in Nature Biotechnology (See our Research Briefing at https://www.nature.com/articles/s41587-024-02203-5) !
References
[1] Sauermann, Henry, and Chiara Franzoni. "Crowd science user contribution patterns and their implications." Proceedings of the national academy of sciences 112.3 (2015): 679-684. https://doi.org/10.1073/pnas.1408907112
[2] Szantner, Attila. "Massively Multiplayer Online Science." Levelling Up: The Cultural Impact of Contemporary Videogames. Brill, 2016. 103-110. https://doi.org/10.1163/9781848884380_012
[3] Sullivan, Devin P., et al. "Deep learning is combined with massive-scale citizen science to improve large-scale image classification." Nature biotechnology 36.9 (2018): 820-828. https://doi.org/10.1038/nbt.4225
[4] Kawrykow, Alexander, et al. "Phylo: a citizen science approach for improving multiple sequence alignment." PloS one 7.3 (2012): e31362. https://doi.org/10.1371/journal.pone.0031362
[5] McDonald, Daniel, et al. "American gut: an open platform for citizen science microbiome research." Msystems 3.3 (2018): 10-1128. https://doi.org/10.1128/msystems.00031-18
[6] Waldispühl, Jérôme, et al. "Leveling up citizen science." Nature Biotechnology 38.10 (2020): 1124-1126. https://doi.org/10.1038/s41587-020-0694-x
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in