Single-cell sequencing edged out other contenders as our choice of Method of the Year in 2013. These techniques really came into their own in 2013 and are fast providing new insights into the workings of single cells that ensemble methods are incapable of.
Back in 2008 we chose next-generation sequencing as our Method of the Year not only because of how the new techniques would improve performance in conventional sequencing applications, but also because they opened up whole new applications, unthinkable with traditional Sanger sequencing. Our choice of Method of the Year in 2013 bears this out, as none of these single-cell sequencing applications would be possible without next-generation sequencing. And in some applications the sequencing is used almost exclusively for identifying and counting tagged molecules.
Our choice likely comes as a surprise to all those who were certain that we would pick CRISPR/Cas9 technology for targeted genome modification. This is certainly an exciting technology, and not only for genome engineering, but also for epigenome editing as described in a Method to Watch. But genome editing with engineered nucleases was our pick for the 2011 Method of the Year and although CRISPR/Cas9 provides a huge practical improvement by largely dispensing with the need to engineer the nuclease and relying instead on a programmable guide RNA, the advance over 2011 is mostly one of ease-of-use.
Methods to investigate biology at the level of single cells have been of keen interest to Nature Methods since the journal started. Our first research article from Robert Singer described a paraffin-embedded tissue FISH (peT-FISH) method to simultaneously detect expression of several genes in situ in single cells while maintaining tissue morphology (Capodieci, P. 2005). This was followed by many other imaging-based methods for such things as measuring cell growth (Groisman, A. 2006), quantifying mRNA (Raj, A. 2008) and protein (Gordon, A. 2006) levels, profiling intracellular signaling (Krutzik, P.O. %26 Nolan, G.P. 2006)(Loo, L.-H. 2007) and DNA insertion-site analysis (Schmidt, M. 2008) in single cells.
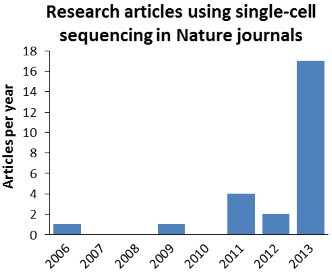
The number of original research articles published in Nature journals exploded in 2013. These numbers may not be complete.
It was only in 2013 that we finally saw an explosion of original research articles using or reporting single-cell sequencing methods in Nature-family journals. Numerous studies reported new biological results that relied on sequencing of whole or partial genomes or transcriptomes from single cells.
Our Method of the Year special feature has three Commentaries by researchers in the field, including some of the earliest developers and users of methods for single-cell analysis. An Editorial, News Feature and Primer describe our choice and provide helpful background information. We hope you enjoy the selection of articles in our special feature.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in