
Written by Pengcheng Shang, Gwen Taylor, Danica Sutherland, and Terence Dermody on behalf of the authors.
Mammalian reoviruses are generalist pathogens with a broad host range in mammals, including humans1. For over three decades, our laboratory has studied reovirus pathogenesis with a focus on identifying host factors governing viral entry. The initial steps in viral infection require interactions with specific cell-surface attachment factors and internalization receptors. These interactions dictate host range, transmission route, tissue tropism, and disease. In newborn mice, reovirus establishes primary infection in the intestine and spreads systemically to sites of secondary infection, including the brain (Fig. 1A)1,2. We previously identified several attachment factors and receptors for reovirus and found that reovirus-host interactions involve discrete virus-receptor pairs that influence pathogenesis at multiple steps (Fig. 1B). However, the identity of proteinaceous receptors that govern reovirus neurotropism remained elusive. In this study, we set out to identify a proteinaceous neuronal receptor for neurotropic T3 reovirus.
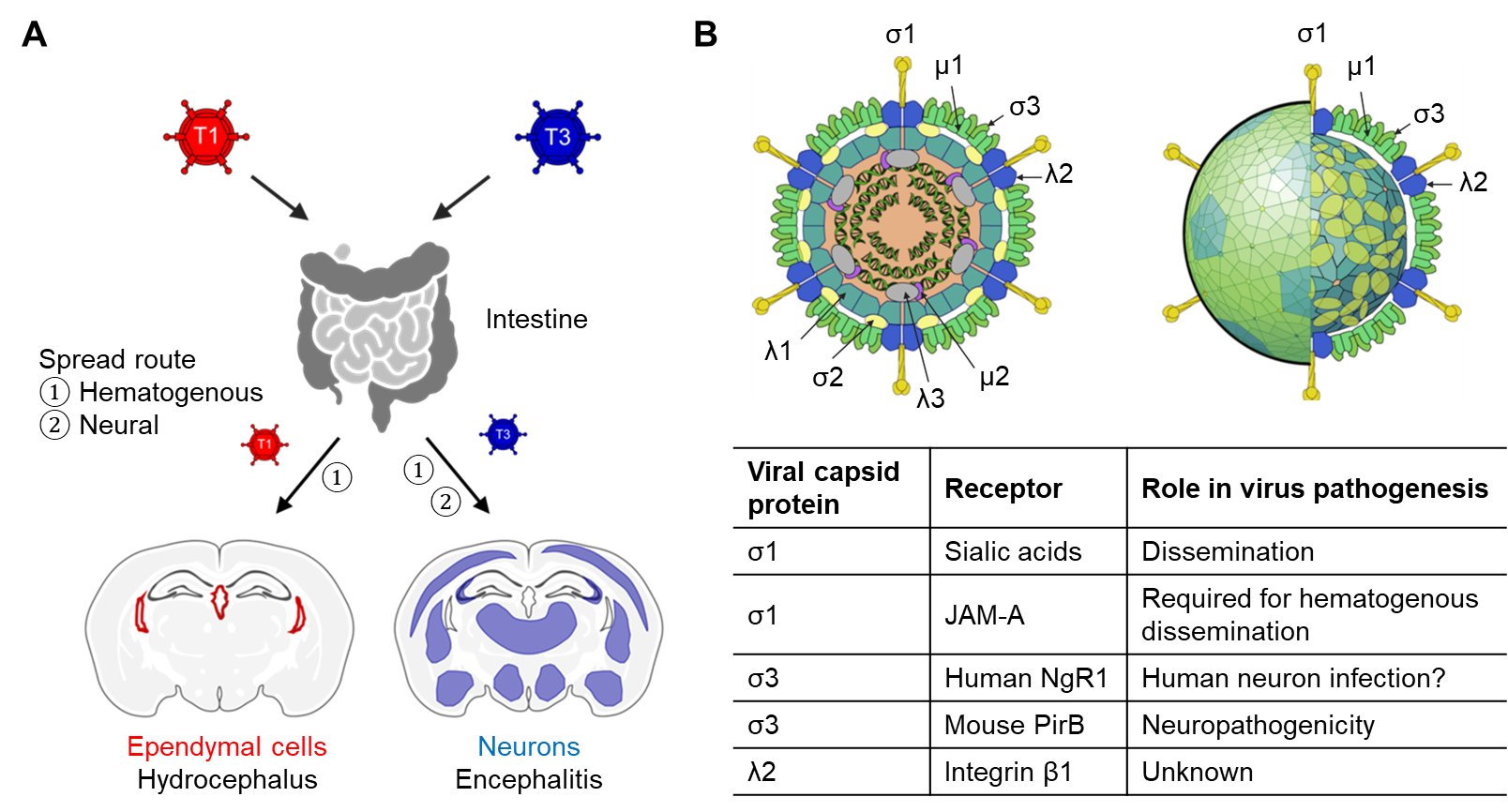
Figure 1. Reovirus pathogenesis and receptors. (A) Reovirus spread and neuropathogenesis in mice is serotype-dependent. Serotype 1, T1; Serotype 3, T3. (B) Reovirus proteins and their host receptors. The reovirus particle images were adapted from ones shown in ViralZone, SIB Swiss Institute of Bioinformatics with permission. Schematic and particle images were prepared using BioRender.
We conducted a gain-of-function CRISPR-activation (CRISPRa) screen to identify receptors conferring reovirus serotype-dependent neuropathogenesis. Unlike loss-of-function screens using CRISPR knockout or siRNA libraries, CRISPRa screens do not require a receptor-expressing cell line and are insensitive to receptor redundancy. We identified paired immunoglobulin-like receptor B (PirB) as the top receptor candidate. PirB is expressed on immune cells and neurons and belongs to the leukocyte immunoglobulin-like receptor (LILR) family, which includes immune-stimulatory LILRA and immune-inhibitory LILRB receptor sub-families3,4. PirB is the sole LILRB expressed in mice, whereas mice express several LILRA orthologs, termed paired immunoglobulin-like receptor A (PirA). PirB also serves as a receptor for major histocompatibility complex I5, myelin-associated inhibitors (MAIs)6, and amyloid-β7. Remarkably, another reovirus receptor, Nogo-66 receptor 1 (NgR1), which we identified using an siRNA library screen8, also binds MAIs.
PirB displays the expected properties of a virus receptor. Ectopically expressed PirB allows reovirus binding to and infection of nonsusceptible cells, and this binding and infectivity is blocked by PirB-specific antibody or recombinant PirB protein. In collaboration with Dr. David Alsteens at the Université catholique de Louvain in Belgium, we defined the kinetics and thermodynamics of the reovirus-PirB interaction using atomic force microscopy (AFM). The application of AFM technology allowed us to assess the biophysics of reovirus binding with receptors coated on model surfaces and expressed on living cells (Fig. 2A)9. We found that reovirus binds PirB with nanomolar affinity and is capable of multivalent interactions. We further found that PirB binds recombinant reovirus capsid protein σ3, which also is engaged by human NgR1, suggesting that σ3 is the reovirus ligand for PirB. The importance of σ3 for reovirus-PirB interactions was additionally highlighted by our finding that reovirus particles depleted of σ3 are incapable of using PirB as a receptor. We now have identified two structurally distinct σ3 receptors, human NgR1 and murine PirB, that also share physiological ligands.
The ectodomains of PirA and PirB share ~ 92% amino acid identity and contain six immunoglobulin (Ig)-like modules (D1-D6)10. However, reovirus preferentially binds PirB. Reciprocal exchanges of PirA and PirB extracellular domain sequences demonstrates that the PirB D3-D4 domains are required for reovirus binding and infection, suggesting flexibility of the PirB extracellular region, as an internal region of the receptor is engaged by the virus (Fig. 2B).
Signaling induced by LILRB receptors, including PirB, is mediated by intracellular immune inhibitory motifs3. Similar to native PirB ligands, reovirus attachment leads to the phosphorylation of these intracellular motifs and initiates PirB intracellular signaling. We hypothesize that immune-inhibitory PirB signaling suppresses host innate immune responses to allow reovirus replication. In addition, PirB signaling regulates actin polymerization by activating cofilin, which subsequently depolymerizes F-actin7. We currently are investigating how F-actin dynamics are regulated by reovirus-triggered receptor signaling for optimal endocytosis efficiency.
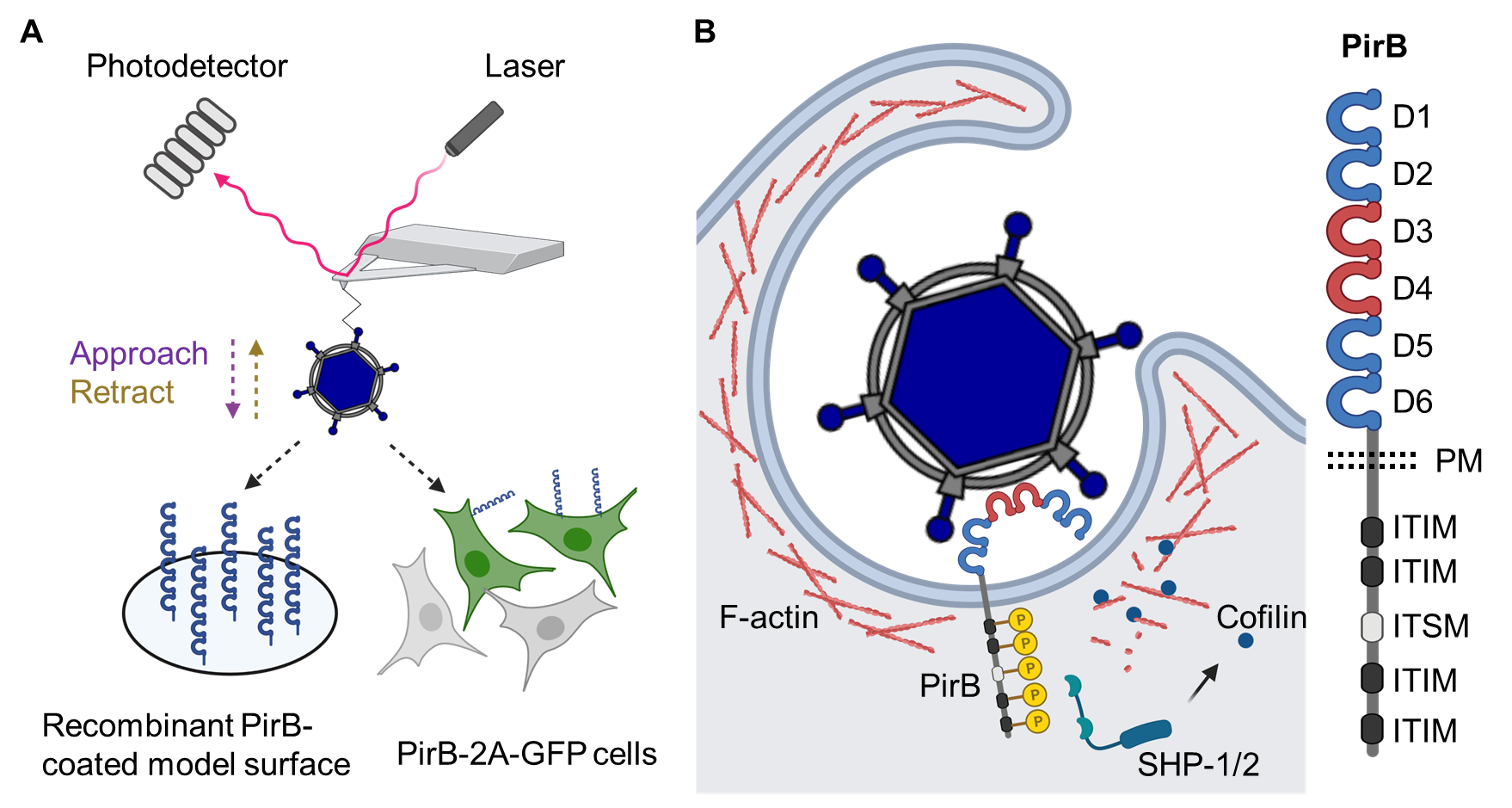
Figure 2. Molecular mechanism of reovirus-PirB interaction. (A) Kinetics and thermodynamics of reovirus-PirB interaction assessed using atomic force microscopy. (B) Model of PirB signaling triggered by reovirus attachment. Schematics were prepared using BioRender.
PirB expressed on neurons contributes to the replication of T3 reovirus in the murine CNS and may influence T3 reovirus neural dissemination. In contrast, PirB is dispensable for hematogenous dissemination and replication of T1 reovirus. Therefore, PirB is a functional neuronal receptor for T3 reovirus. We hypothesize three key mechanistic roles for PirB in reovirus infection of neurons: (i) PirB may coordinate reovirus macropinocytosis at axonal termini and contribute to the retrograde transport of reovirus in endosomal vesicles11; (ii) Axon-localized PirB may promote reovirus attachment to non-myelinated axons and allow lateral motility to sites of endocytosis12; and (iii) PirB may facilitate reovirus spread across synapses to promote dissemination via neuronal circuits. The precise roles of PirB in reovirus entry will be defined in our future research.
Mechanisms regulating viral entry into host cells have been one of the most significant and intriguing topics in the field of virology. The blueprint of reovirus entry discovered in our research highlights the value of reovirus as a tractable experimental model to investigate how viruses co-opt multiple host receptors in attachment and internalization and how the highly coordinated entry process influences viral tropism and disease.
Reference
1 Dermody, T. S., Parker, J. S. & Sherry, B. in Fields Virology Vol. 2 (eds D. M. Knipe & P. M. Howley) 1304-1346 (Lippincott Williams & Wilkins, 2013).
2 Sutherland, D. M., Aravamudhan, P. & Dermody, T. S. An orchestra of reovirus receptors: Still searching for the conductor. Advances in Virus Research 100, 223-246 (2018).
3 Burshtyn, D. N. & Morcos, C. The expanding spectrum of ligands for leukocyte Ig-like receptors. J Immunol 196, 947-955 (2016).
4 van der Touw, W., Chen, H. M., Pan, P. Y. & Chen, S. H. LILRB receptor-mediated regulation of myeloid cell maturation and function. Cancer Immunol Immunother 66, 1079-1087 (2017).
5 Takai, T. Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology 115, 433-440 (2005).
6 Atwal, J. K. et al. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science 322, 967-970 (2008).
7 Kim, T. et al. Human LilrB2 is a β-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer's model. Science 341, 1399-1404 (2013).
8 Konopka-Anstadt, J. L. et al. The Nogo receptor NgR1 mediates infection by mammalian reovirus. Cell Host Microbe 15, 681-691 (2014).
9 Koehler, M. et al. Glycan-mediated enhancement of reovirus receptor binding. Nature Communications 10, 4460 (2019).
10 Vlieg, H. C., Huizinga, E. G. & Janssen, B. J. C. Structure and flexibility of the extracellular region of the PirB receptor. J Biol Chem 294, 4634-4643 (2019).
11 Aravamudhan, P. et al. Reovirus uses macropinocytosis-mediated entry and fast axonal transport to infect neurons. PLoS Pathog 16, e1008380 (2020).
12 Syken, J., Grandpre, T., Kanold, P. O. & Shatz, C. J. PirB restricts ocular-dominance plasticity in visual cortex. Science 313, 1795-1800 (2006).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in