The Quest for Evolutionary Control
Published in Ecology & Evolution
A holy grail in current evolutionary biology research is the development of a technique for preventing pathogens and cancer from evolving resistance to therapy. Currently, we imagine such a technique would use pre-computed treatment regimens to precisely guide the evolutionary trajectory of a population. The implications of achieving this level of fine grained control would be profound. In addition to the aforementioned drug resistance problem in cancers and infectious diseases alike, fine-grained evolutionary control could give us power over agricultural ecosystems, the gut microbiome, and even larger scale ecosystems.
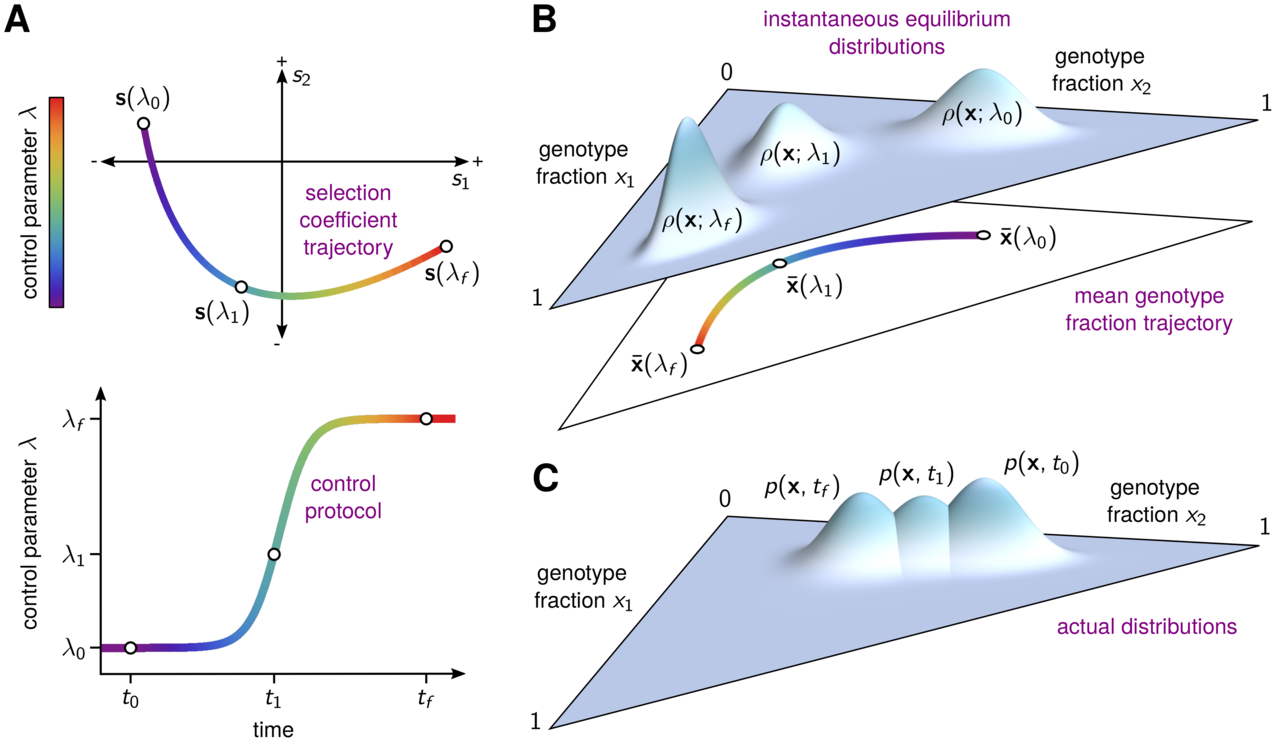
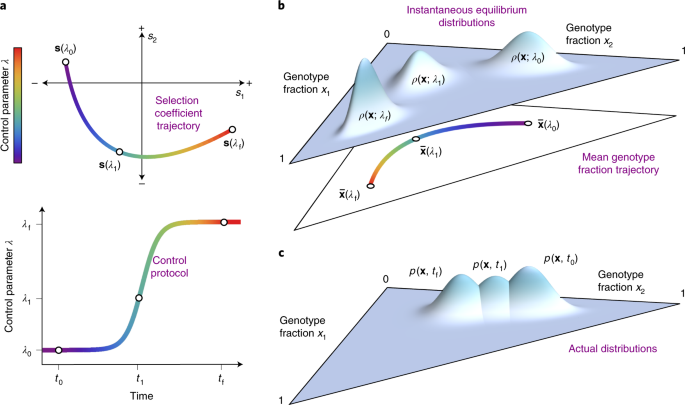
We can think of treatment protocols as sequences of fitness landscapes that we subject a population to. By manipulating these protocols, we can push the population around a local space of possible genotypes. Every new axis of environmental change gives us an additional control knob to use for this purpose. Our hope is that by turning these knobs in the correct way and in the correct sequence, we can push the population to a region of the fitness landscape where it can be driven to extinction or trapped in a valley that will allow us control in the long term.

Encouragingly, we have previously shown mathematically that our evolutionary steering theory is superior, on average, to an arbitrary ordering of drugs and feasable for heterogenous cell populations with dynamic drug responses. Furthermore, in bacteria as well as multiple cancer types, our group has experimentally identified drug pairs or sequences that can potentially be used to steer a population away from resistance.
However, our evolutionary steering approach has serious practical limitations. First, the protocol had to be applied for arbitrarily long times to allow the population to reach a quasi-equilibrium. How long is long enough?
Secondly our initial theoretical work assumed the entire population to be genetically and thus phenotypically homogenous. Our subsequent experimental work demonstrated that heterogeneity and variation between evolutionary replicates further limits the applicability of evolutionary steering.
In real life scenarios, we generally cannot afford to wait around for long enough after every change in drug dosage to be certain that the target population has reached a desired equilibrium. Patient’s can’t wait asymptotically long times. Consequently, even if we knew an approximate sequence of treatments that would guide the population to a desired configuration, we wouldn't know how long we needed to wait after each step in the protocol before moving on to the next.
In our most recent paper, we set out to solve this timing problem. Our solution hinges on an idea borrowed from a radically different field - the concept of counterdiabatic (CD) or transitionless driving in quantum physics. CD driving involves control and manipulation of states evolving in finite time, and has been applied in contexts such as quantum computing and optical traps for ultra cold atoms. CD driving allows us to move a population around on a dynamic, time varying fitness landscape (seascape) as if it were equilibrating instantaneously to each new environment. In reality, we achieve this by applying a different sequence of environments, specified by our explicitly computed CD driving protocol.
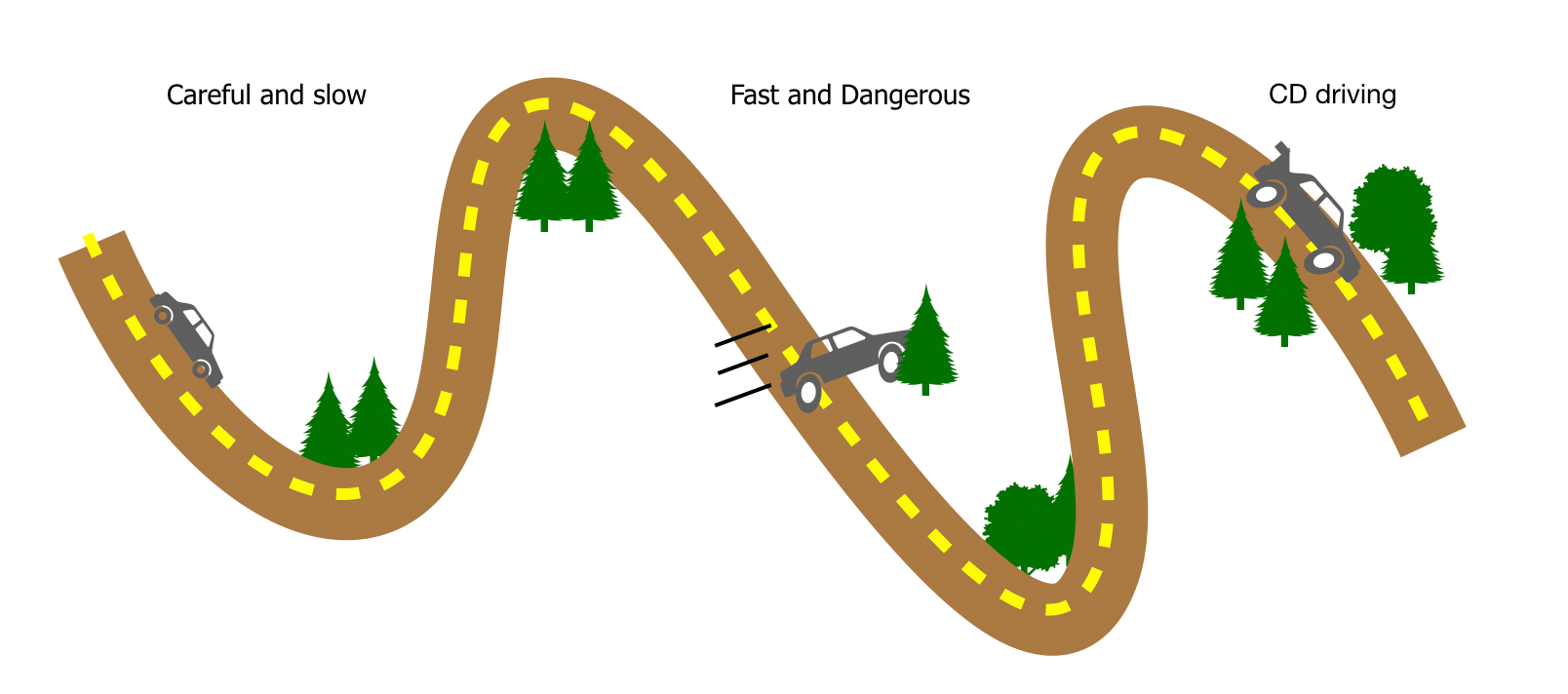
To understand the essence of CD driving, it is helpful to consider a more familiar scenario. Imagine driving a car along a winding road through the mountains, with many sharp curves. In this analogy each point along the road corresponds to a different distribution of mutants in the cellular population (or different quantum states in the original incarnation of the idea). The goal is to get from the start point to the final destination in a short time, but always staying on the chosen road. Veering off-road can be dangerous: for example, you may want to avoid certain mutants along the way that can lead to complications for the patient. The route poses no problems for slow drivers: by gradually accelerating, breaking, and steering, one can reliably guide the car through even the most perilous hairpin. In biological terms, slow driving would be a gradual protocol of changing drug dosage, with the mutant distribution adapting in response, keeping on target throughout. But attempting the same trip at high speeds is risky, since you will lose traction going around sharp turns and swerve off the road. Analogously, simply speeding up the original drug protocol will lead the cell population to stray from the target distribution of mutants. The solution is to figure out a strategy that counteracts the effects of high speeds. For the car this might be adding spoilers or wings to the back, pushing the car downwards and increasing grip during fast turns. Ideally, the spoilers may even dynamically adjust to optimize control on the fly. Similarly, CD driving provides a dynamic correction force in the form of altered dosage or even adding new types of drugs into the mix, with the aim of achieving the desired distribution of mutants at each moment of time.
We developed a general method to calculate and implement this CD driving strategy using information about the fitness of the different types of mutants in the population. To illustrate and validate our CD driving prescription in silico, we tried them out in empirically-derived fitness landscapes for genes conferring resistance to the anti-malarial drugs pyrimethamine and cycloguanil. Normally, when we increase the concentration of one of these drugs, the population takes quite a while to reach equilibrium. When we use CD driving, however, the population stays approximately at equilibrium the entire time. This behavior is illustrated in the video below, where each vertex represents a genotype, lines between vertices indicate mutational adjacency, the size of the red circles indicates the frequency of each genotype we would expect to see at equilibrium, and the size of the blue circles indicates the frequency of each genotype that we observed at each point in time. There are error bars, too, but they're mostly too small to see. Notice that without CD driving there is a substantial lag between the observed genotype distribution and the equilibrium distribution. CD driving nearly eliminates this lag.
This is just the beginning of an exciting new direction applying ideas from quantum physics to evolutionary biology, what we’re calling quantum-inspired evolutionary biology. Additionally our work is at the forefront of the burgeoning field of evolutionary therapy - using our understanding of evolution to design novel therapies for cancer as well as infectious diseases. To this end, we have a lot of work to do now in identifying the circumstances under which driving will and will not work and testing these ideas in the wet lab. Getting this far required assembling a large team of physicists, evolutionary biologists, and computer scientists. We are looking forward to seeing what other fields we need to bring together to achieve our next steps in CD driving for evolving populations, as well as quantum inspired evolutionary biology and evolutionary therapy at large.
Follow the Topic
-
Nature Physics

This journal publishes papers of the highest quality and significance in all areas of physics, pure and applied.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in