The South American Microbiome Archive
Published in Microbiology and Protocols & Methods
In the last years we have witnessed the emergence of associations between the human gut microbiome and multiple health conditions, which are often world-region specific. Therefore, an important challenge in microbiome research is to adequately characterise human populations across the world and particularly in currently neglected locations. This would likely allow regional idiosyncrasies in microbiome variability to be distinguished from robust global signatures of gut microbiome influences over human health, a pressing challenge for the development of new microbiome-based biomedical strategies.
However, current efforts to characterise the gut microbiome are skewed towards high income countries. More than 70% of the public human microbiome data comes from samples taken from people in Europe or North America, although their combined population represents less than 15% of the global population. This disparity effectively means that entire continents rely on results from research conducted in wealthier countries that may not face the same diseases they struggle with, and whose main findings may fail to be generalized to their populations.
Towards that end, new global and local efforts are collecting samples from previously neglected populations. However, these efforts often involve enrolling large human cohorts with diverse lifestyles, which increases the costs and can make them prohibitively expensive in world regions with more restricted scientific budgets. Thus, other strategies to understand global microbiome patterns are required.
Recently, the largest collection of public human microbiome data analysed under a unified bioinformatics analysis was released. The resource, called the Human Microbiome Compendium (HMC) is an effective way to systematically collate our knowledge about global microbiome diversity. Importantly, this initiative highlighted a worrisome pattern: the least studied world regions, such as South America, are among the most biodiverse.
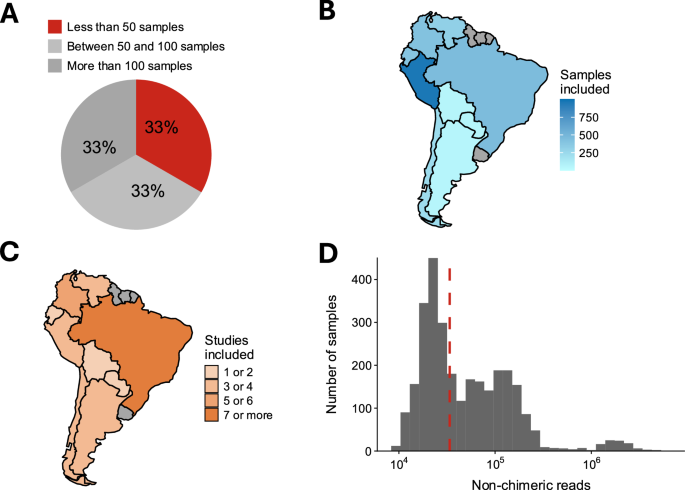
Although the resource is already impressive, we identified two aspects where we could contribute further value to it. First, the software used in the creation of the HMC is not available to the public yet. Second, some decisions made while building the HMC likely exacerbates existing differences in microbiome characterisation efforts across the globe. For instance, the criteria to only include studies with more than 50 samples.
Thus, we downloaded the HMC and reanalysed it to assess how we could contribute. The exploration showed that world-regions with lower proportion of high-income countries tend to include less samples than wealthier continents. Moreover, we identified that the original world-region used (Latin America and the Caribbean) could be subdivided for finer-grain resolution, as people living in South and Central America show distinct microbiome signatures. Finally, I come from Chile, so I had a longstanding interest and passion about advancing South American microbiome studies. Thus, we set to expand the known universe of South American gut microbiomes, a region among the most biodiverse yet extremely understudied.
What are the key findings?
We conducted a search across public repositories of microbiome data, and identified more than 3,000 samples coming from 9 out of the 13 South American countries. Interestingly, approximately 73% of the identified studies weren’t included in the HMC, and a third was just because of the exclusion of small primary microbiome studies. We built a simplified replica of the software used in the HMC and used it to reanalyse all samples we identified. The resulting collection is the South American MicroBiome Archive: saMBA.
After reanalysing the samples, we identified more than 1,300 new genera not previously described in the gut microbiomes of South Americans. Moreover, this expanded collection of studies allowed us to better characterise the diversity across the continent, describing it as the second most biodiverse on earth, slightly below Central America. However, we also identified that despite these new efforts, the regional biodiversity is still underestimated, further highlighting the need of taking new samples from South American individuals.
We recognised that scientific budget in the continent may be more limited compared to other regions, so we performed a continental characterisation of the current sampling effort to maximise the scientific return of the money invested in taking new samples. First, we determined which countries are the most biodiverse by comparing all its samples. Then, we identified what countries are lagging in terms of their representation, calculated by using the number of samples of the country compared to the share of the total population in the continent. This allowed us to identify countries where the need for new samples is more pressing.
Thus, we believe our research highlights the real extent of the continental biodiversity, while helping prioritising locations where to conduct new sampling efforts. We suspected our experience further charactering the South American continent may be shared with other world regions where the HMC didn’t fully capture their biodiversity. Because we believe the HMC is such an important resource and other world regions may benefit from expanding their own collection of microbiomes, we made our software, which is compatible with the pipeline used in the HMC, available to other researchers. We hope this can propel microbiome research globally.
What comes next?
We are interested in expanding saMBA to include other body sites and whole genome sequencing data. The HMC clearly showed that Latin America and the Caribbean is the world region with the largest proportion of unknown taxas. Thus, expanding the catalogue of known genomes from the continent is a major challenge in the microbiome field that has potential to open new areas of research.
Ecologically, it is thought provoking that the world region with highest biodiversity in terms of multicellular organisms also has the highest diversity in terms of gut bacterial commensals. The region seems to be a hot spot of world’s biodiversity across all levels of organismal complexity.
Regarding biomedical research, better understanding the different compositions of gut microbiomes could expand our understanding of what a healthy microbiome looks like. This, although disguised as a simple question, is the source of active debate in the scientific community and evidences that despite the significant progress made in the last decade, there is still much more work to do. Thus, further characterising the gut microbiome of different world regions is a promising avenue to disentangle local idiosyncrasies from global patterns of how the microbiome influences our health. This has unparallel potential to identify biomarkers of disease across global populations, and holds promise to open new therapeutic approaches.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in