The speciation continuum below your feet
Published in Ecology & Evolution, Microbiology, and Genetics & Genomics

Earth soils are inhabited by millions of species of bacteria, archaea, protists, animals, and plants. We focused on the cyanobacterium Microcoleus, which is among the most abundant primary producers in soil and soil crusts across the globe. While this name probably does not ring a bell, everyone has likely encountered green slime on wet soil in their garden or in a puddle in fields. This is a Microcoleus mat. Microcoleus forms filaments that combine into bundles by their polysaccharide sheaths, which stick the filaments together. Microcoleus inhabits all continents and latitudes. It thrives in wet and temperate soils as well as in the most extreme environments, such as hot and cold deserts and dry Himalayan high-altitude valleys. It can cover bare soil as a slime, but it also forms so-called soil crusts by aggregating the soil particles with polysaccharides, making a more stable environment for other organisms. It has been estimated that the soil crusts cover more than 12% of the terrestrial environments.

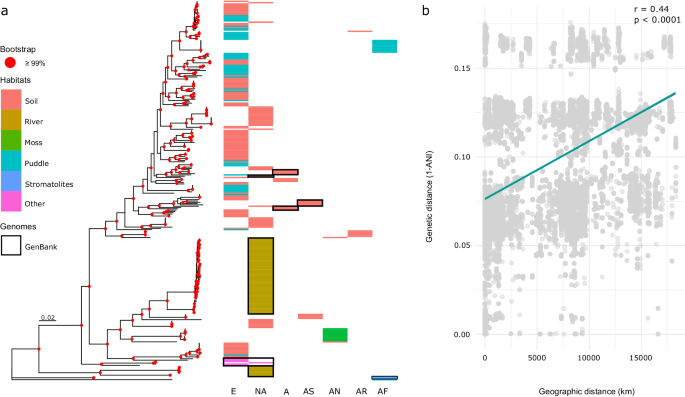
The processes leading to species emergence – speciation, are still far from understood. This gap is even more pronounced in microbes due to their small dimensions and incredible species diversity, reaching billions of species. We chose the cyanobacterium Microcoleus to delve deeper into unraveling this intriguing biological mystery. We walked, ran, and biked thousands of kilometers to gather hundreds of soil samples. We isolated almost 500 strains of Microcoleus and sequenced more than 200 genomes, including eight herbarium specimens. Herbarium specimens were sequenced in the Natural History Museum London under the ‘supervision’ of Charles Darwin’s statue, which stands in the main hall of the museum.
After curating the genome dataset, we started with the reconstruction of the evolutionary relationships. Applying several phylogenetic approaches, we discovered that Microcoleus encompasses remarkable genome diversity, comprising at least 12 lineages that diverged 5-15 million years ago during the Pliocene and Miocene epochs. Then, we started to investigate the factors shaping the lineage divergence. First, we asked if geographical distance plays a role in the diversification. It was expected that microbes have unlimited dispersal abilities and that the local environmental conditions select the species. However, these assumptions were based on morphological data, which are known to hinder genomic diversity. We found that genetic and geographical distance were correlated, which suggests that the physical distance shapes the diversification in Microcoleus. As we mentioned, Microcoleus inhabits harsh conditions; therefore, we expected the environmental conditions to play a significant role in the diversification. We obtained numerous environmental variables for our dataset. We found that the climate and soil properties drive the diversification by several independent tests. Furthermore, we identified several dozens of genes that seemed to be under the selection of environmental factors. The genes were mostly involved in stress response to the environmental conditions.
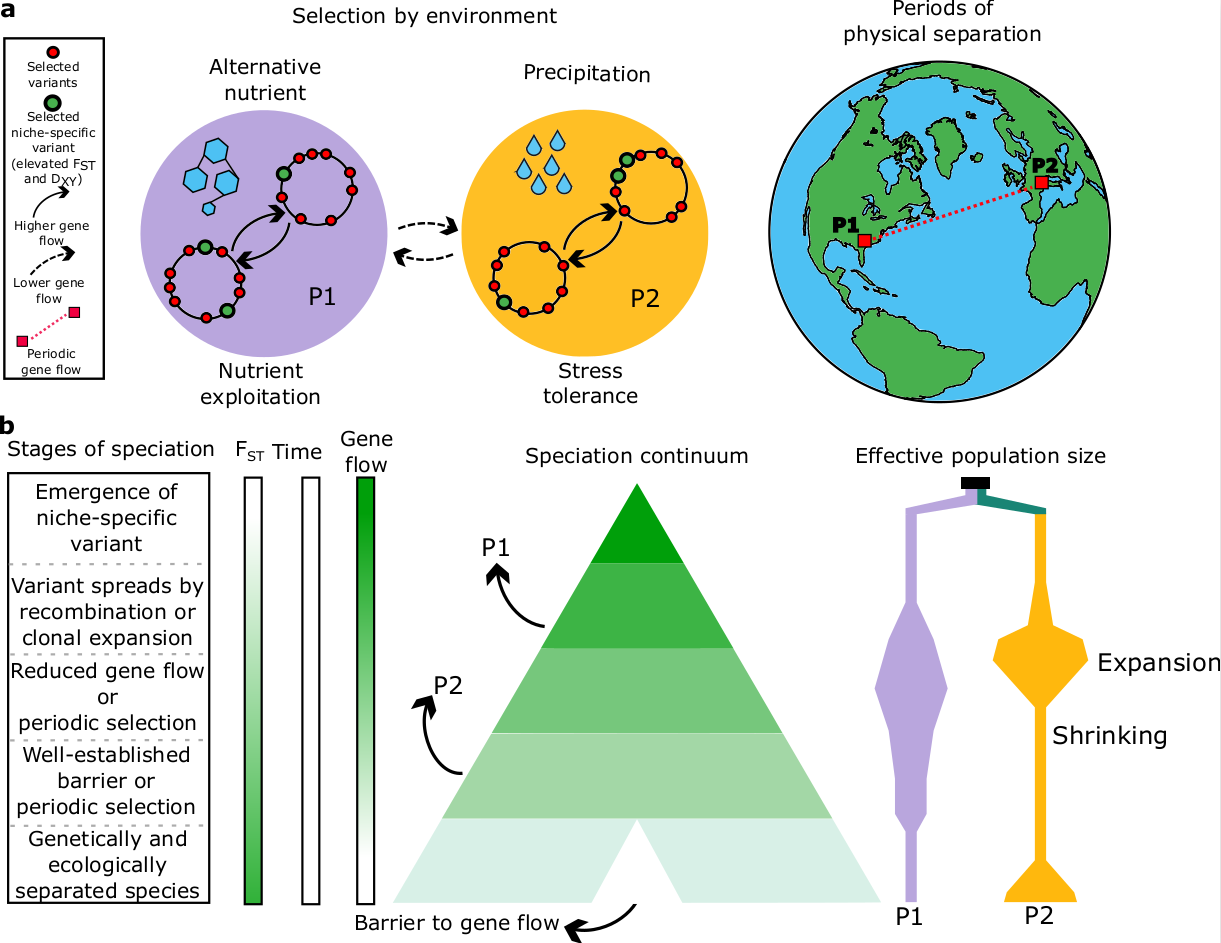
All prokaryotes, including cyanobacteria, have asexual reproduction; therefore, they are considered clonal. However, an extensive body of evidence shows that the prokaryotic genome is shaped by gene flow in a similar way to plants and animals. We observed a spectrum of gene flow barriers among Microcoleus lineages, ranging from tens of percent of the genome affected by gene flow to instances where gene flow was nearly absent. This made us think: what other organisms show similar patterns of speciation with gene flow? Since prokaryotes are still severely understudied with respect to the evolutionary gene flow patterns, we turned to the realm of animals. Stankowki and Ravinet proposed a definition of a speciation continuum, where the diverging lineages are separated by diverse strengths of reproductive isolation. Cyanobacteria lack sexual reproduction, but they have intensive gene flow. We extended the speciation continuum to include diverging lineages with varying strengths of gene flow and observed all stages of diversification in Microcoleus, from minimal divergence with intensive gene flow to complete divergence without gene flow. This makes Microcoleus a unique study system for the speciation continuum.
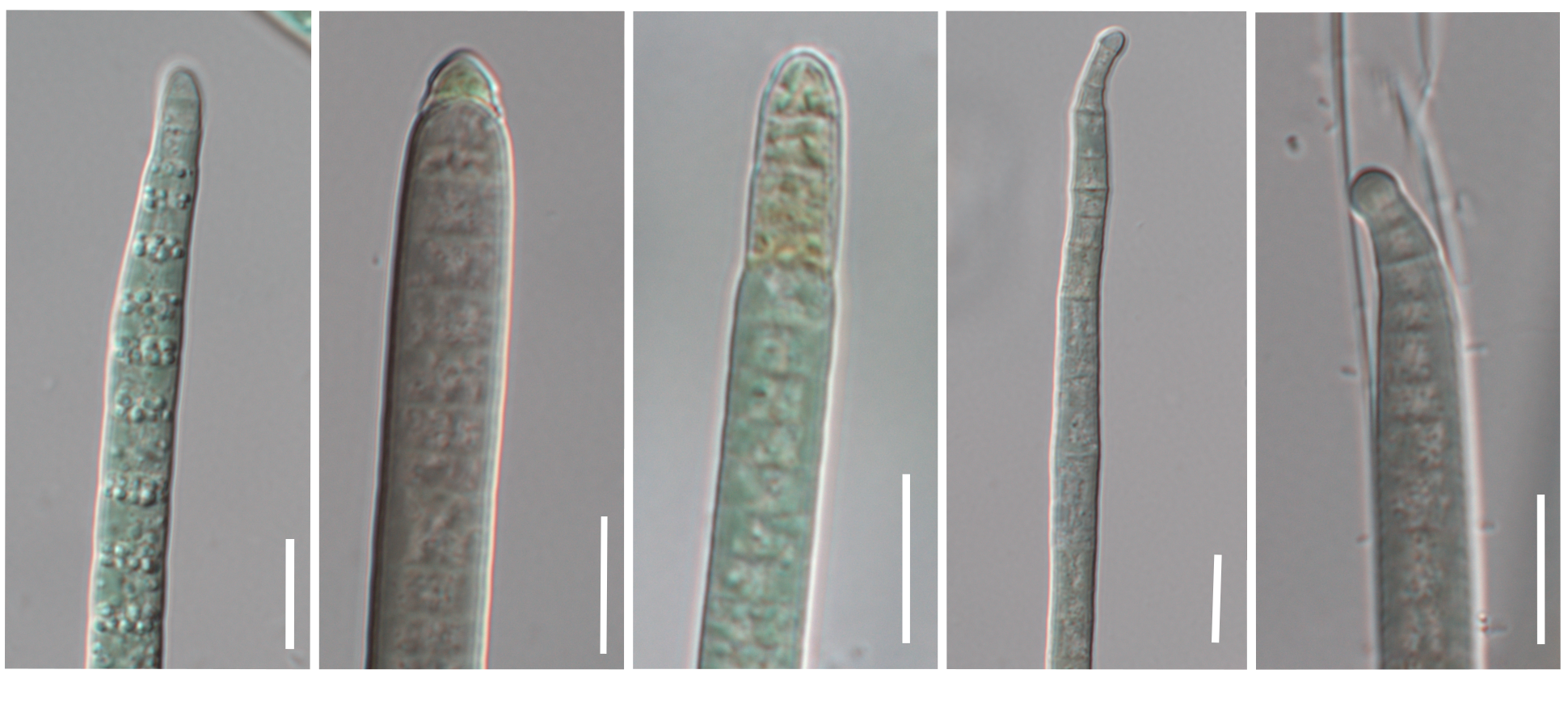
Cyanobacteria are rather unique among the prokaryotes because they have extensive morphological variability. We tested a connection between genomic and morphological diversification in the speciation continuum. Microcoleus has one unique feature – the filaments are attenuated towards the ends, and they have a little hat called a calyptra. Its function has not been rigorously tested, but it is expected to have a protective function. The filaments are moving, not only on the soil surface, but by boring in the soil as well. We noticed that different lineages have distinct shapes of the filaments apices. Some lineages have shorter, some longer, some thicker or more narrow apices. To study this further, we developed a measure of the filament pointiness. We found that the pointiness diversification follows the genome diversification. We could clearly recognize most of the lineages from others, but three were indistinguishable. Furthermore, we investigated a hypothesis of whether the filament pointiness relates to several environmental variables, which could suggest its importance in diversification. We found that the filament pointiness significantly correlates with soil properties variables, suggesting that the shape of the apex may have an adaptive function.
The microbial world below our feet is immensely diverse and plays a key role on a global scale in terrestrial ecosystems and biochemical cycles. This hidden world is evolving and ever-changing. From now on, whether you are jogging or gardening, we hope you can appreciate a little bit more the green cyanobacterial rug on the soil or in a puddle. It is a population of evolving speciation continuum of Microcoleus.
Written by Petr Dvořák, Aleksandar Stanojković, Svatopluk Skoupý
The research was supported by the Grant Agency of the Czech Republic (grant no. 19-12994Y and 23-06507S).
References:
Stanojković A, Skoupý S, Johannesson H, Dvořák P. The global speciation continuum of the cyanobacterium Microcoleus. Nat Commun. 2024; 15(1):2122. doi: 10.1038/s41467-024-46459-6
Skoupý S, Stanojković A, Casamatta DA, McGovern C, Martinović A, Jaskowiec J, Konderlová M, Dodoková V, Mikesková P, Jahodářová E, Jungblut AD, van Schalkwyk H, Dvořák P. Population genomics and morphological data bridge the centuries of cyanobacterial taxonomy along the continuum of Microcoleus species. iScience. 2024; 27(4):109444. doi: 10.1016/j.isci.2024.109444
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in