The structure of the bacterial surface exclusion protein TraT
Published in Microbiology and Cell & Molecular Biology

Bacterial conjugation: a driver of antibiotic resistance
Antibiotic resistance is one of the greatest public health challenges that can be conferred by a variety of mechanisms. One such mechanism, bacterial conjugation, involves a plasmid-encoded multiprotein complex spanning the bacterial cell envelope, known as the type IV secretion system.1 This machinery enables bacteria to pass on resistance plasmids to each other, increasing the number of bacteria resistant to antibiotics.
To this day, there are still may unanswered questions about bacterial conjugation. The molecular basis of an important stage of conjugation, surface exclusion, remains to be elucidated. Surface exclusion is the final stage of conjugation that prevents the transfer of identical or closely related plasmids into a bacterium that already carries them.2 The main goal of my PhD was to focus on the type IV secretion component that is involved in surface exclusion, the outer membrane lipoprotein TraT.3 Thirty-five years ago, preliminary electron microscopy studies reported TraT to be a ‘multisubunit, doughnut-shaped’ protein with a ‘central region'.4 However, no further structural studies were conducted by the authors. As a structural biologist, this was a fascinating observation. If TraT truly is a doughnut-shaped protein, how does it mediate the exclusion of bacterial plasmids if it resembles a pore? There was only one way to get closer to the answer, and that was to firstly start by solving the structure of TraT.
Structural elucidation of TraT
As many people in the field of structural biology know, cryo-electron microscopy (cryo-EM) has developed significantly over the past two decades, surpassing the limitations of other structural techniques like X-ray crystallography, which I firstly used in an attempt to solve the structure of TraT. Unfortunately, I only obtained TraT crystals that produced very low resolution diffraction. With cryo-EM being an effective method for determining the structures of large, multisubunit proteins, it was decided that the structure of TraT would best be solved by this technique. This was an exciting opportunity to work with cutting-edge technology, where I learned how to prepare cryo-EM samples and how to process cryo-EM data.
Obtaining the structure of TraT was not an easy process; I encountered many issues during my attempts to (1) express TraT in bacteria, (2) purify TraT from bacteria, and (3) generate optimum cryo-EM grid conditions for TraT. However, after optimization, monodisperse particles of TraT were visualized on a Titan Krios, revealing ‘doughnut-like’ particles that, upon closer inspection, resembled a champagne bottle cork. This was a euphoric moment, as this observation aligned with the one made 35 years ago.

revealed TraT to resemble a champagne bottle
cork
Following data collection and optimization of image processing, the structures of two TraT homologs from Escherichia coli and Klebsiella pneumoniae were solved. They were both found to form a structurally conserved decamer with subtle amino acid differences. Interestingly, they were found not to form a channel in the bacterial outer membrane but were instead anchored to the outer membrane via a diacylglycerol and palmitic acid-modified α-helical barrel domain.
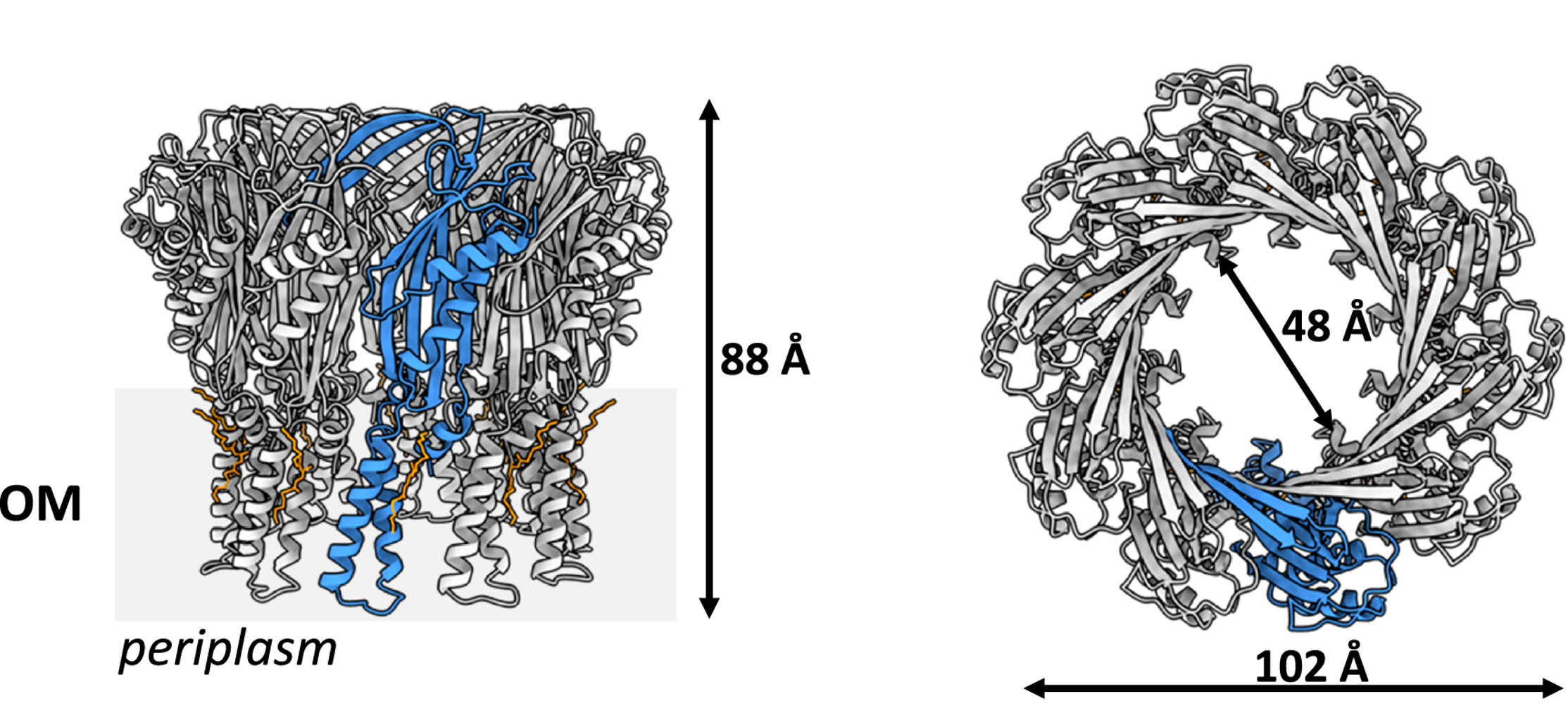
champagne bottle cork-like structure and is anchored to the outer membrane.
(OM = outer membrane; a single protomer is coloured blue)
Additionally, my colleagues revealed through bioinformatic analyses that TraT emerged as a chromosomal gene before being incorporated into diverse plasmid families. From a conjugation point of view, our findings pose many questions and theories about how TraT can prevent plasmid transfer into a bacterium. Could it be that TraT acts as a decoy, or does it have a binding partner through which it executes surface exclusion with? These questions are yet to be answered. The TraT structure will undoubtedly aid in solving this intriguing mechanism and help to better understand the mechanism of conjugation, a mediator of antibiotic resistance.
References
- Lawle TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. Jul 15 2003;224(1):1-15. doi:10.1016/S0378-1097(03)00430-0
- Kingsman A, Willetts N. The requirements for conjugal DNA synthesis in the donor strain during flac transfer. J Mol Biol. Jul 5 1978;122(3):287-300. doi:10.1016/0022-2836(78)90191-2
- Achtman M, Kennedy N, Skurray R. Cell--cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc Natl Acad Sci U S A. Nov 1977;74(11):5104-8. doi:10.1073/pnas.74.11.5104
- Sukupolvi S, O'Connor CD. TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiol Rev. Dec 1990;54(4):331-41. doi:10.1128/mr.54.4.331-341.1990
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in