Therapeutic phages that infect mycobacteria require cell-surface trehalose polyphleates
Published in Microbiology
Written by K. S. Wetzel on behalf of all authors
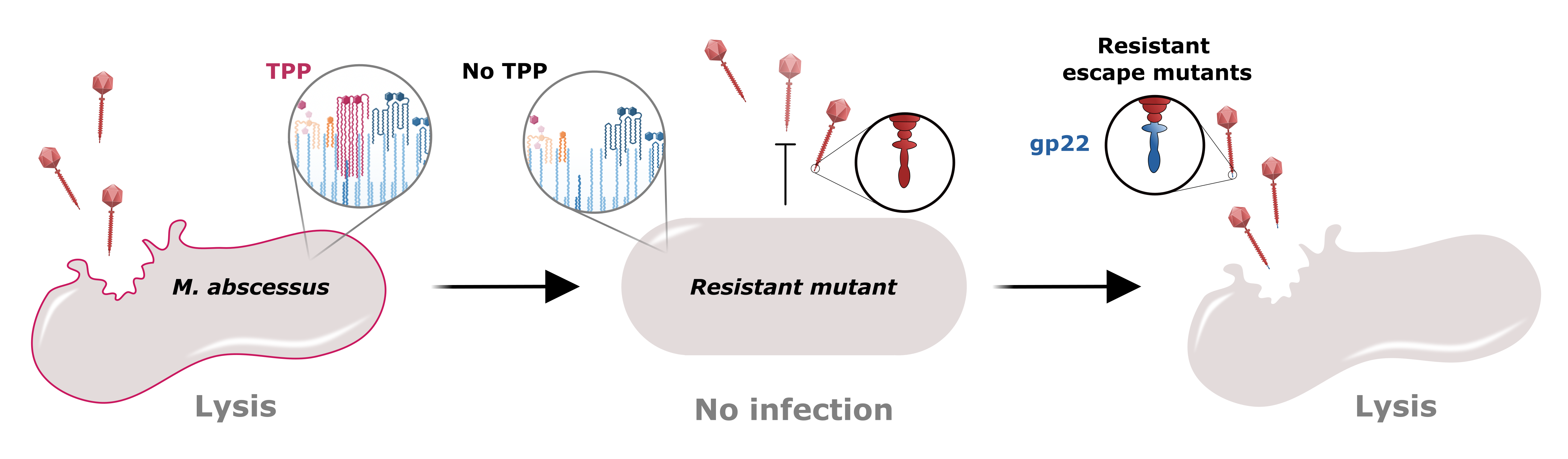
The rise of infections by antibiotic resistant bacteria are a major public health concern. Non-tuberculous mycobacteria (NTM) are a group of such bacteria and include the opportunistic pathogen Mycobacterium abscessus. NTM infections are broadly antibiotic resistant and prolonged use of antibiotics against them is often poorly tolerated by patients. These infections are common among people with cystic fibrosis and other obstructive pulmonary diseases and can disqualify them from lung transplants. Therefore, alternate therapies are greatly needed.
One such alternative are phages, viruses that specifically infect and kill bacteria. Phage therapy was first proposed over a century ago but was not pursued in western countries once antibiotics were discovered. However, interest in phage therapy has grown in response to the rise of antibiotic resistant bacteria. As such, in 2017, the Hatfull lab (Pittsburgh, USA) collaborated with physicians of a pediatric cystic fibrosis patient in the UK who had a disseminated M. abscessus infection that could not be cleared with antibiotic treatment. The Hatfull lab has an extensive collection of phages isolated on a harmless cousin of M. abscessus, Mycobacterium smegmatis. These phages have been isolated by undergraduate students all over the world as part of the SEA-PHAGES authentic research course. Out of therapeutic options, doctors asked if phages in the Hatfull lab’s collection could infect and kill this patient’s strain.
Extensive screening efforts identified several phages that infect this strain (M. abscessus GD01) and a cocktail of three phages was prepared and administered intravenously. The three phages were chosen because they are genetically distinct from each other, minimizing the likelihood that the bacteria would develop resistance to two or more phages. The patient improved and was able to return to her daily life, although the infection was not entirely resolved. This report prompted a steady influx of patient strains to the Hatfull lab for screening for phage susceptibility. However, we quickly learned that many patients’ M. abscessus strains are not susceptible to these same phages, such that any treatment must be personalized. Additionally, many strains are not susceptible to any phages tested, and more phages are clearly needed. Furthermore, we do not know how NTM strains become resistant to phages, and what mechanisms could cause co-resistance to several phages. Broadening phage therapy to more NTM patients requires a better understanding of the determinants of phage susceptibility and pathways to resistance.
To address these questions, I set out -together with other members of the Hatfull lab, Christian Chalut in Toulouse (France), and Morgane Illouz, Silke Malmsheimer, and Laurent Kremer in Montpellier (France)- to determine which host factors are required for phage infection of M. abscessus GD01. We created a library of GD01 cells where each cell had a random mutation in the form of a transposon insertion. Then, we infected the library with phage, and recovered the survivors; these survivors had an insertion in a gene required for phage infection. To create the library, we first needed to re-engineer phage that delivers the transposon, as the transposon was marked with kanamycin resistance to allow selection of successfully mutated cells. Many M. abscessus strains – including GD01– are not sensitive to kanamycin. Using our previously reported CRISPY-BRED engineering systems, I constructed a hygromycin-resistant transposon for use in M. abscessus.
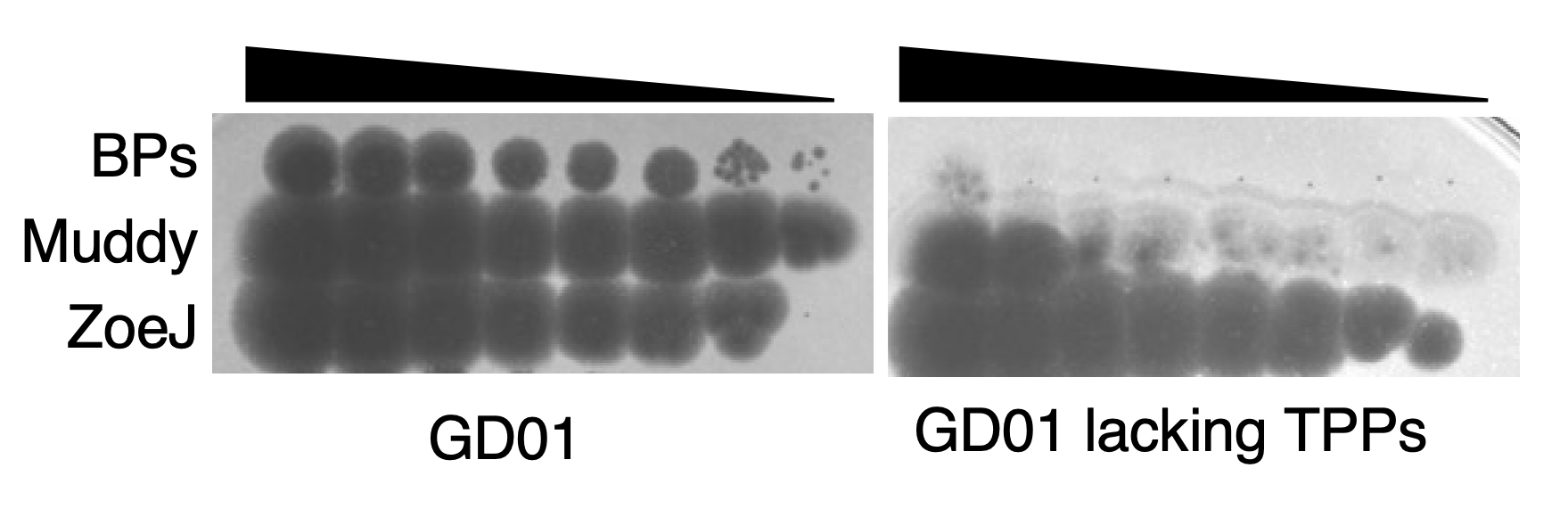
We infected the GD01 library with two phages that were used to treat the patient; BPs∆33HTH_HRM10 (hereafter called BPs) and Muddy. Since these two phages are not related genetically, we expected GD01 cells that survived BPs to have different mutations than those that survived Muddy. Surprisingly, all of the mutants for both phages had transposon insertions in genes belonging to the same pathway! This pathway is for the synthesis of trehalose polyphleates (TPPs), very large molecules located at the surface of many species of mycobacteria. These glycolipids were originally described in Mycobacterium phlei decades ago but more recently have been characterized by Christian Chalut’s lab. However, the biological functions of TPPs are not known. Morgane Illouz showed that TPPs are absent from these mutants. We observed that other phages (such as ZoeJ) that infect GD01 do not require TPPs for infection.
While I was identifying GD01 insertion mutants resistant to BPs or Muddy, Haley Aull isolated a series of spontaneously occurring M. abscessus mutants that were resistant to BPs or Muddy. Excitingly, some of these mutants also had mutations in TPP pathway genes! Then, the Hatfull and Kremer labs collaborated to figure out the mechanism for phage resistance.
We found that phage BPs is not able to attach to or kill cells lacking TPPs and that BPs mutants that have small changes in a protein predicted to interact with the cell are able to overcome this resistance mechanism. Lawrence Abad analyzed our large collection of M. abscessus clinical isolates and, together with Madison Cristinziano, showed that several strains have large deletions or insertions in TPP pathway genes that confer phage resistance. We also found that these phages required TPPs to infect the model organism M. smegmatis; this will allow future studies to occur in a biological system that is easier to manipulate. Moreover, use of this organism allowed us to show that some phages other than BPs or Muddy also use TPPs to infect and kill the bacteria.
Lastly, we infected the same GD01 library with the versions of BPs and Muddy that do not require TPPs for infection. These phages killed all of the TPP-deficient mutants that we identified in the first screen, and unveiled mutants that have insertions in different genes required by the phages. Excitingly, none of these new mutants are resistant to both BPs and Muddy.
In summary, this study identified TPP loss as a pathway to phage resistance for the therapeutic phages BPs and Muddy. This co-resistance was unexpected, as these phages are genetically unrelated and do not always infect the same strains. It emphasizes the need to better understand how phages and their host cells interact and the possible pathways to phage resistance. This study also revealed a strategy to circumvent this co-resistance; TPP-independent mutants of BPs and Muddy infect and kill TPP-deficient cells, and we did not isolate a strain resistant to both phages. Therefore, a M. abscessus strain would be much less likely to develop resistance against a phage cocktail containing both TPP-independent versions of BPs and Muddy. As such, these TPP-independent phage versions expand our collection of phages and are now being used therapeutically.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in