In 2005, being a group leader at the MPI-CBG in Dresden, I planned a lecture on lipids for the institute’s annual PhD-course. I looked for original experimental data that would visualize the flow of fatty acid through the Kennedy pathway, but I failed to find it. So I went to the lab and did a pulse-chase experiment with radiolabeled oleic acid in A431 cells. By TLC separation and X-ray film detection I obtained the picture shown below. It nicely shows the flow of the labeled fatty acid, it has a good signal-to-noise ratio and a very good time resolution, but it lacks lipid species resolution, it is limited to the major lipid classes and is at best semi-quantitative. Experts also see the overload in the DAG and TAG spots since a million cells was necessary to obtain enough signal.
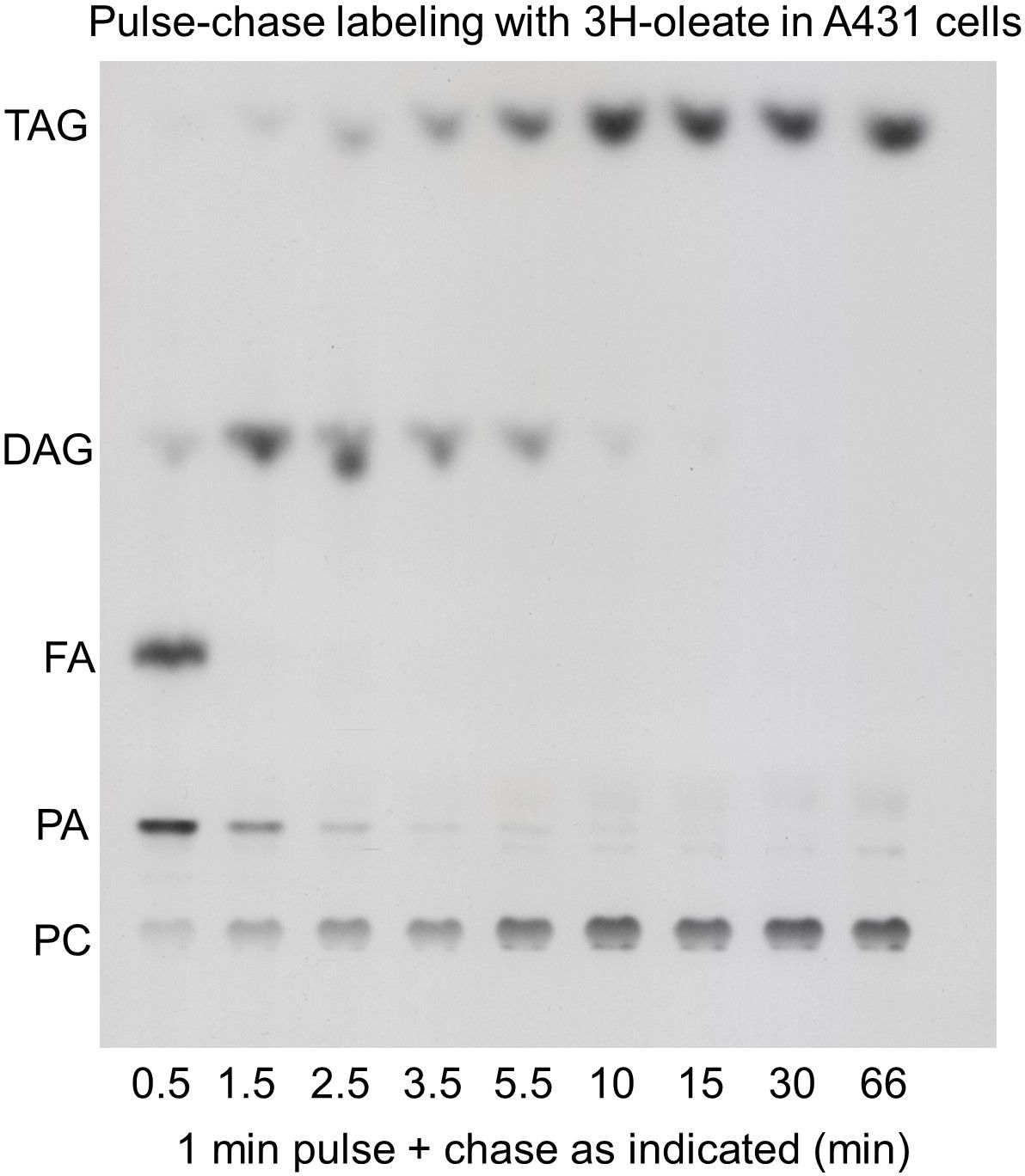
In the absence of better alternatives, we nonetheless continued to use similar labeling experiments to trace the metabolism in parallel to microscopy with fluorescent pentaene-lipids (Kuerschner et al. 2005). In 2009 we moved from Dresden to the University of Bonn into a brand-new institute. Unfortunately, the isotope lab was 100 m away in the neighboring building and the frequency of labeling experiments dropped drastically. At the same time, click chemistry had become popular. To solve the labeling crisis we developed a system based on alkyne lipids and click reaction to a fluorescent dye (Thiele et al. 2012). Still combined with TLC analysis, the sensitivity was much better, quantification was easier and the hotlab became unnecessary. However, lipid species analysis was still out of reach and minor classes like ceramides or lysolipids were hard to see in the background of the total. Anyway, already since the late 1990s mass spectrometry was the dominating technology in lipidomics of unlabeled samples, and a logical next step was to use MS for tracing, in particular since we obtained an Orbitrap MS in 2017.
Why not simply use deuterium- or 13C-labeled fatty acids for tracing? The answer lies in the complexity of lipid meatbolism. As seen above, fatty acid metabolism takes place on the minute scale, so it needs labeling times in the minute range to resolve it kinetically. Still, hundreds of labeled species are produced in these short pulses, most of them in very small amounts. Now imagine you have a palmitic acid with four deuterium atoms as the label. At a very realistic degree of labeling of 1% you obtain the following if you look at one of the major triglycerides:
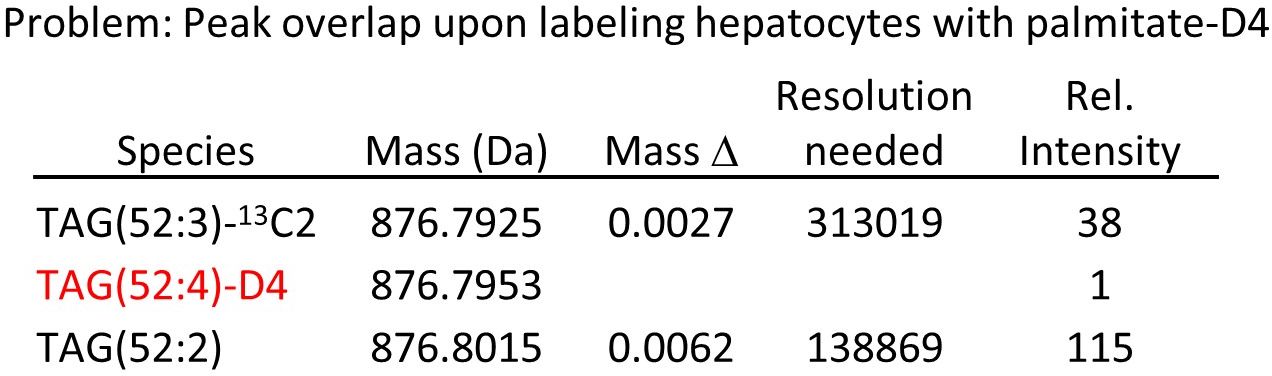
Your labeled peak falls in between two much stronger unlabeled peaks at distances of 3 and 6 mDa, very unlikely to be resolved even by the latest generation of Orbitrap machines. The situation improves a bit if you allow more D or 13C in the label, but then another problem arises: by complete or partial degradation, the labeled atoms will distribute into other fatty acids and give secondary labeling, and this will result in practically untractable complexity.
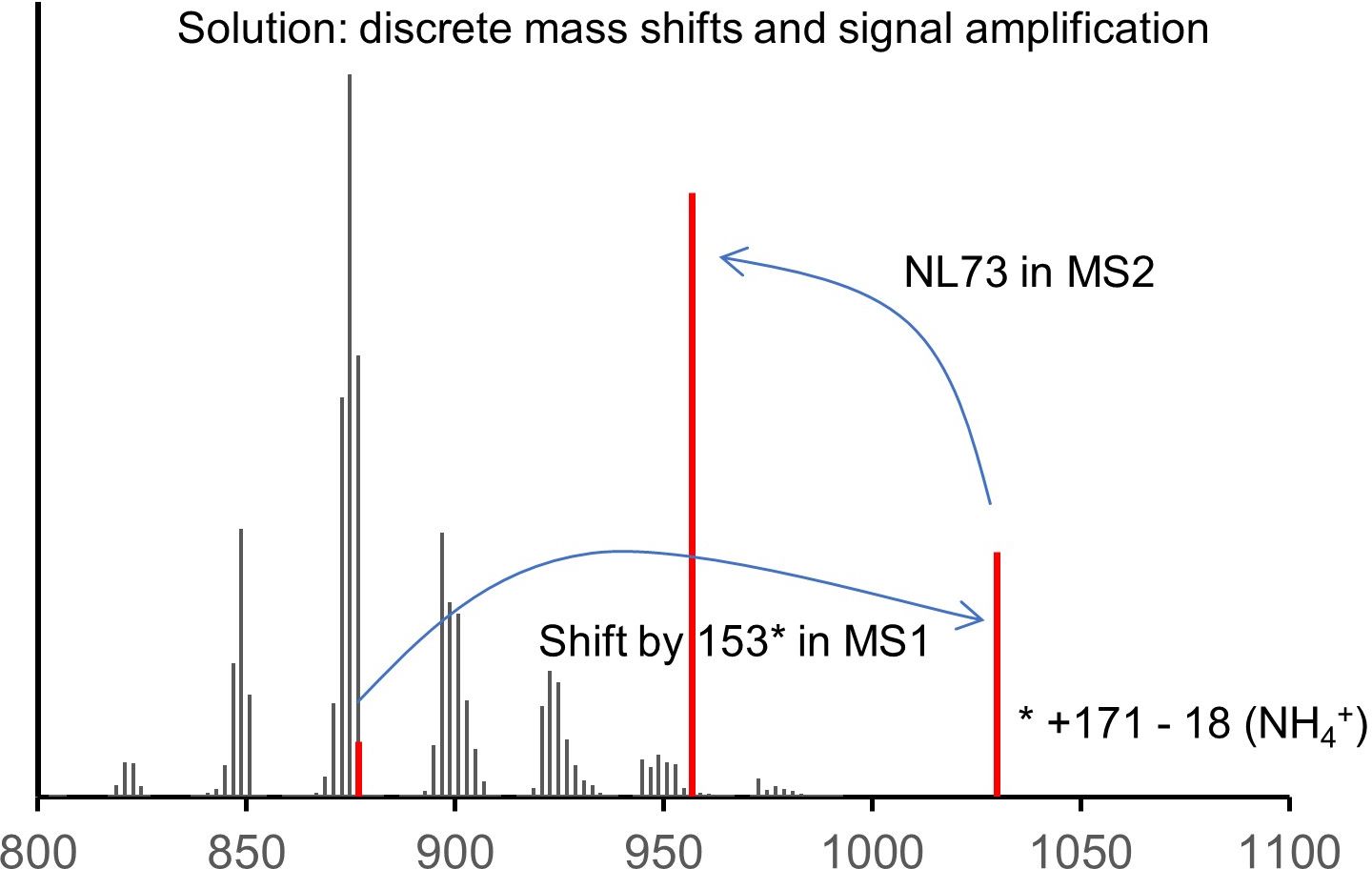
In the new method, the solution to that is quite simple. The triple bond serves as a unique feature discriminating the label from the rest. The click reporter C171 translates the presence of the triple bond into a large mass shift in MS1 and a unique, predictable fragmentation in MS2, and it amplifies the signal by its permanent charge. The structure of the molecule allows for a multiplex approach, saving time and making data better comparable.
What is the potential of the method? I hope it will make lipid metabolism better accessible – for those who are genuinely interested in it, and for those to whom it is just a side aspect. The former will like the time resolution and sensitivity, the latter should benefit from flexibility and robustness. It should enable to study the complexity of fatty acid metabolism in whole animals as well as rapidly testing drug candidates for effects in lipid metabolism. And of course, the system as such might still be improved: more internal standards will make more classes quantifiable, and multiplexing could be combined with multi-labeling, both further increasing the density of data. Lipid tracing will become an omics-type of experiment and the increase in information will need a professional data evaluation infrastructure. But still, it should be fun: see the hundreds of compounds that are derived from a single labeled fatty acid and admire the perfect regulation that keeps such a system functioning and under control
Kuerschner, Lars; Ejsing, Christer S.; Ekroos, Kim; Shevchenko, Andrej; Anderson, Kurt I.; Thiele, Christoph (2005): Polyene-lipids: a new tool to image lipids. In Nature methods 2 (1), pp. 39–45. DOI: 10.1038/nmeth728.
Thiele, Christoph; Papan, Cyrus; Hoelper, Dominik; Kusserow, Kalina; Gaebler, Anne; Schoene, Mario et al. (2012): Tracing fatty acid metabolism by click chemistry. In ACS chemical biology 7 (12), pp. 2004–2011. DOI: 10.1021/cb300414v.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in