Transcriptomics in cancer revealed by Positron Emission Tomography radiomics
Published in Cancer
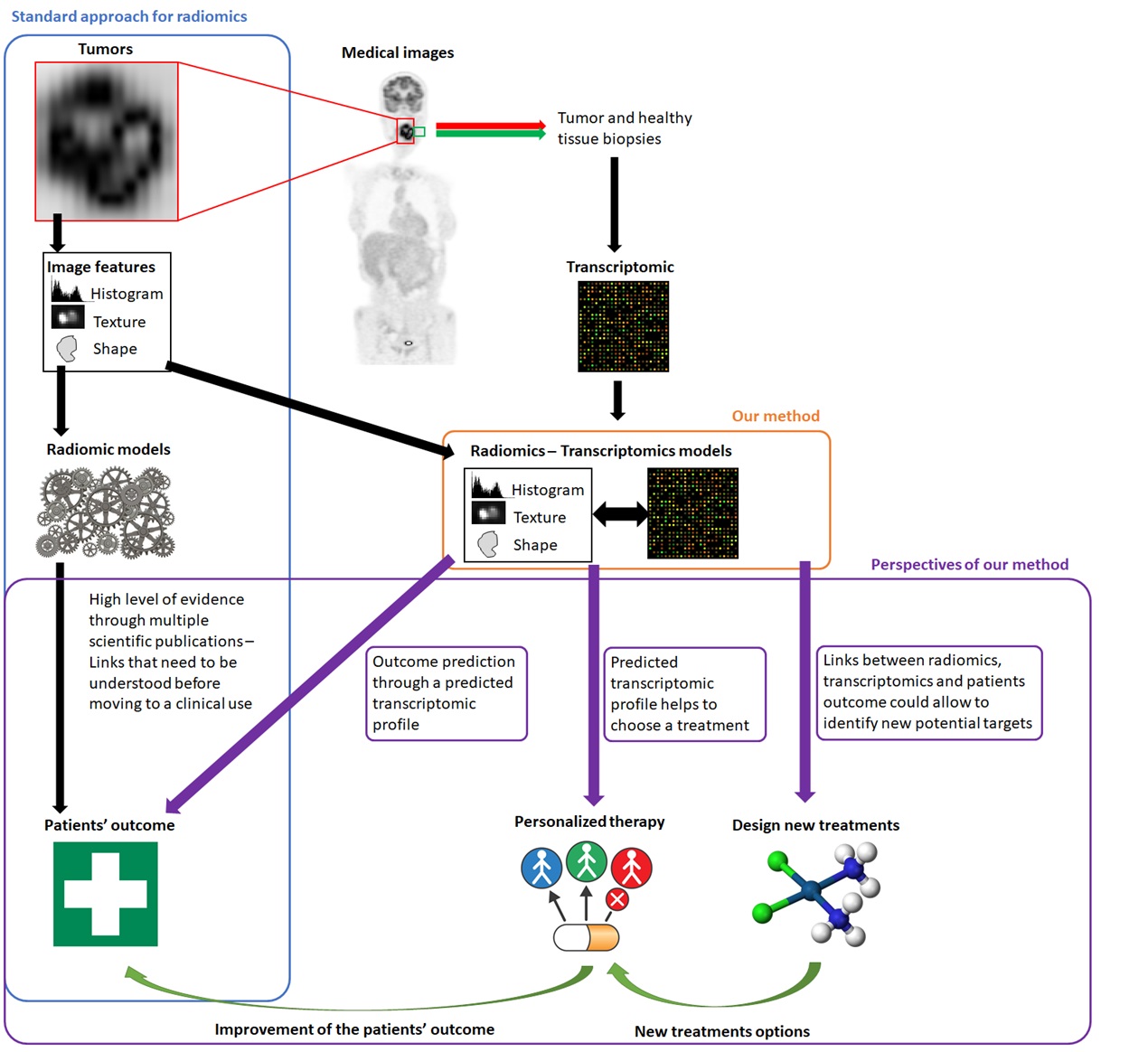
Radiomics denotes the high-throughput extraction of a large number of quantitative measurements (called handcrafted or engineered image features) from any kind of medical images. These features have been shown useful to identify disease characteristics (e.g., clinical stage, molecular subtype, etc.) or to predict patients’ outcome (e.g., response to therapy, survival, etc.) for many cancer types.
The underlying assumption, and the rationale behind the development of the radiomics approach for medical imaging, have always been that these image features reflect, at least partly, the underlying physiological processes, e.g. glucose metabolism, angiogenesis, etc., which could explain their predictive and prognostic values observed in different cohorts of patients. Within this context, we decided, in 2012, to start the prospective recruitment of head and neck cancer patients undergoing both 18F-FDG positron emission tomography (PET) imaging and transcriptome-wide gene expression analysis in the cancerous tissue, in order to determine to what extent PET image radiomics could indeed reflect different gene pathways’ activities. We found such associations between radiomic features from PET images and gene modules, and thus with biological pathways specifically altered in the cancer tissue, as compared to healthy tissue.
The interest for radiomics has raised exponentially (from 2 publications in 2012 up to more than 700 in 2019, according to Web of Science), with a strong increase in the confidence level of the clinical value of radiomics, thanks to improvements in methodology and use of increasingly larger cohorts of patients. However, the limited ability of clinicians to translate radiomic features into biological processes is a major limitation for integration in routine clinical practice. Indeed, radiomic features can capture details in images that the naked eye, even the expert one, cannot necessarily appraise. Hence, our results help understanding the radiomics prognostic and predictive value observed in previous studies. They also strengthen the value of radiomics as a promising approach to personalize treatments through targeting tumor-specific molecular processes. We can foresee the possibility, that routinely acquired medical images may ideally complement a clinical evaluation with radiomics-based interpretation of the underlying activity of gene expression pathways. This should in turn allow to select the most appropriate therapy treatment as well as to predict outcome. The method we developed has several advantages. Firstly, we performed the analyses with no a priori consideration of the gene expression patterns from previous knowledge. Consequently, this approach could allow discovering potential new targets for the design of innovative treatments. Secondly, the approach provides a biological meaning for the radiomic signatures, which may in turn allow moving a step further towards the potential use of radiomics in clinics.
- Florent Tixier, Research Fellow, LaTIM, INSERM UMR1101, Univ Brest, Brest, France
- Mathieu Hatt, Junior Researcher, LaTIM, INSERM UMR1101, Univ Brest, Brest, France
- Catherine Cheze-Le-Rest, Professor, Department of Nuclear Medicine, Poitiers University Hospital, Poitiers, France
- Laurent Corcos, Director of Research, INSERM, UMR 1078, Université de Brest, Génétique Génomique Fonctionnelle et Biotechnologies, Etablissement Français du Sang, Brest, France
- Dimitris Visvikis, Director of Research, LaTIM, INSERM UMR1101, Univ Brest, Brest, France
Follow the Topic
-
Scientific Reports

An open access journal publishing original research from across all areas of the natural sciences, psychology, medicine and engineering.
Related Collections
With Collections, you can get published faster and increase your visibility.
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026
Women’s Health
Publishing Model: Open Access
Deadline: Feb 28, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in