Trimming glycans to make most of biomedical samples. Sweet and sensitive CE-MS method for quantitative characterization of native N-glycomes: from ng-level blood isolates to single cells
Published in Chemistry
In the Ivanov laboratory at Northeastern University, our research is focused on the development of innovative sample preparation and nanoflow-based liquid phase separation techniques coupled with mass spectrometry for molecular (e.g., proteomic, glycomic) profiling of amount-limited biological and biomedically-relevant samples. By minimizing the number of sample preparation steps and employing more efficient, low volume (nL-/pL-level)-based analytical techniques, we aim to detect, identify, structurally characterize, and quantify more molecular features (e.g., proteins and glycans) at lower sample amounts compared to conventional techniques.
Recently, we developed a highly sensitive capillary electrophoresis-mass spectrometry (CE-MS) method for N-glycan profiling of complex and limited biological samples (e.g., human blood isolates) using the APTS tag to improve the separation and detectability of released N-glycans. The developed glycan labeling-based approach resulted in the detection of >200, >400, and >500 N-glycans in isolates of total serum IgG, plasma-derived extracellular vesicles (EVs), and total plasma, respectively, for injected sample amounts equivalent to <500 nL of blood 1. Yet, we observed that the yield of APTS-labeling was less than 60%, and that some N-glycans remained mostly or completely unlabeled, especially highly sialylated ones. Nevertheless, such highly negatively charged glycans could be detected in their native non-labeled state, resulting in increased N-glycan coverage in the three types of blood-derived isolates. Based on this observation, we aimed to develop an alternative strategy, where the glycans would be analyzed in their native underivatized state 2. The developed label-free CE-MS method resulted in the detection of >250, >310, and >520 N-glycans in isolates of total serum IgG, plasma-derived EVs, and total plasma, using injected amounts equivalent to <25 ng of serum proteins and nL-levels of plasma-derived samples. Compared to previously reported results for biological samples of similar amounts and complexity, the number of identified N-glycans was increased up to ~15-fold, and unmatched profiling sensitivity was achieved for sub-0.2 nL of plasma volume equivalents. Also, previously undetected highly sialylated (up to 10 SiA residues) N-glycans, which could represent a new class of clinical biomarkers, were identified and structurally characterized.
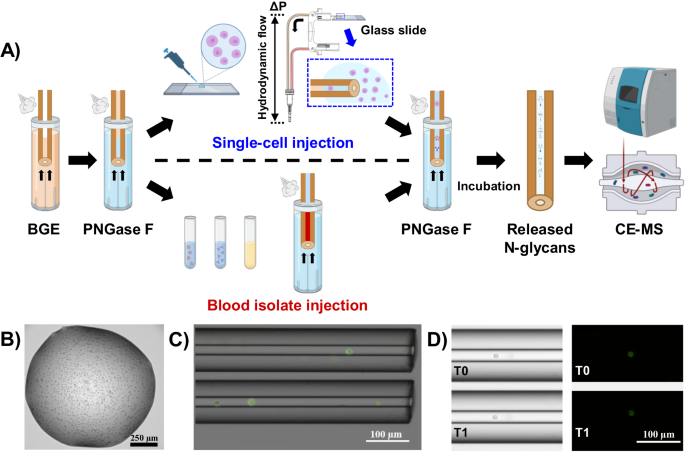
In the study presented here, we pushed the boundaries of detection sensitivity and demonstrated the sensitivity levels sufficient to profile the N-glycome of single cells with the implementation of an in-capillary sample preparation method coupled online to the previously developed label-free CE-MS approach 2. The analytical workflow we recently reported for in-capillary sample processing and CE-MS-based top-down proteomic profiling of small populations of cells and single cells 3 was adapted to the profiling of N-glycans released from minute amounts of biological samples as well as single cells. Fig. 1 depicts the analytical workflow we developed for N-glycan profiling of single cells and ng-levels or pL-levels of blood isolates (e.g., IgM, IgG, EVs, and total plasma) or other types of amount-limited biomedically/clinically-relevant specimens.
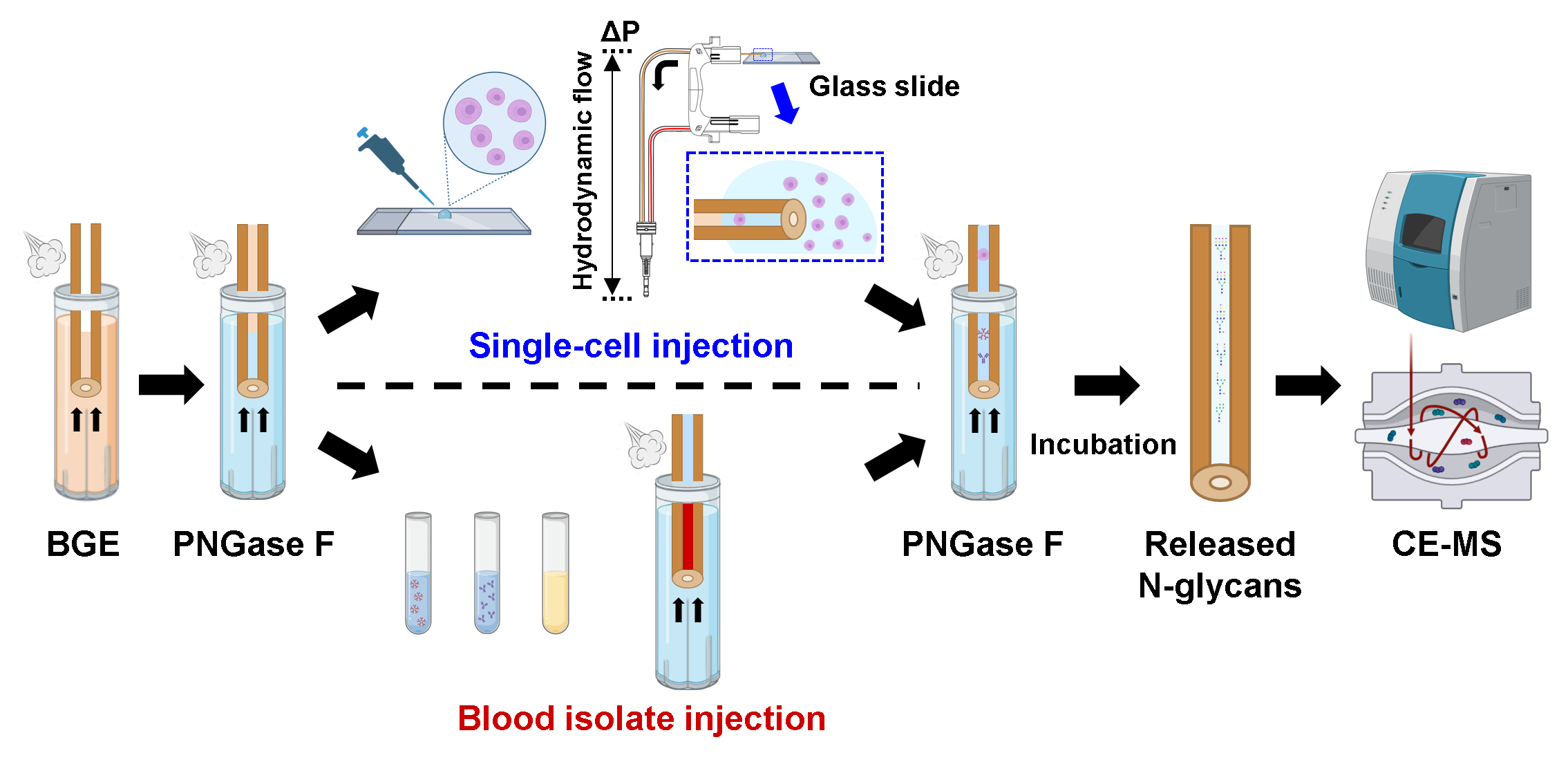 Fig. 1: Analytical workflow for in-capillary sample processing coupled to label-free high-sensitivity CE-MS for native N-glycan profiling of single cells and amount-limited biomedically-relevant samples (e.g., blood-derived isolates). The injected samples are sandwiched between two plugs of PNGase F and incubated inside the capillary. After the digestion step, the CE and ESI-MS voltages are triggered for online CE-MS analysis of the released N-glycans in their native non-labeled state.
Fig. 1: Analytical workflow for in-capillary sample processing coupled to label-free high-sensitivity CE-MS for native N-glycan profiling of single cells and amount-limited biomedically-relevant samples (e.g., blood-derived isolates). The injected samples are sandwiched between two plugs of PNGase F and incubated inside the capillary. After the digestion step, the CE and ESI-MS voltages are triggered for online CE-MS analysis of the released N-glycans in their native non-labeled state.
Our proof-of-concept experiments showed that the developed CE-MS-based workflow could result in the detection, separation, identification, and quantitation of up to 100 N-glycans in one single mammalian cell (Fig. 2). Specific N-glycosylation patterns were demonstrated for HeLa and U87 single cells. Interestingly, N-glycome alterations were observed at the single-cell level when HeLa and U87 cells were stimulated with lipopolysaccharide (LPS) (Fig. 2). The mild deglycosylation conditions allowed us to preserve the cell integrity and predominantly release intact and native cell surface N-glycans, which may potentially benefit the multi-omic and spatial characterization of individual cells.
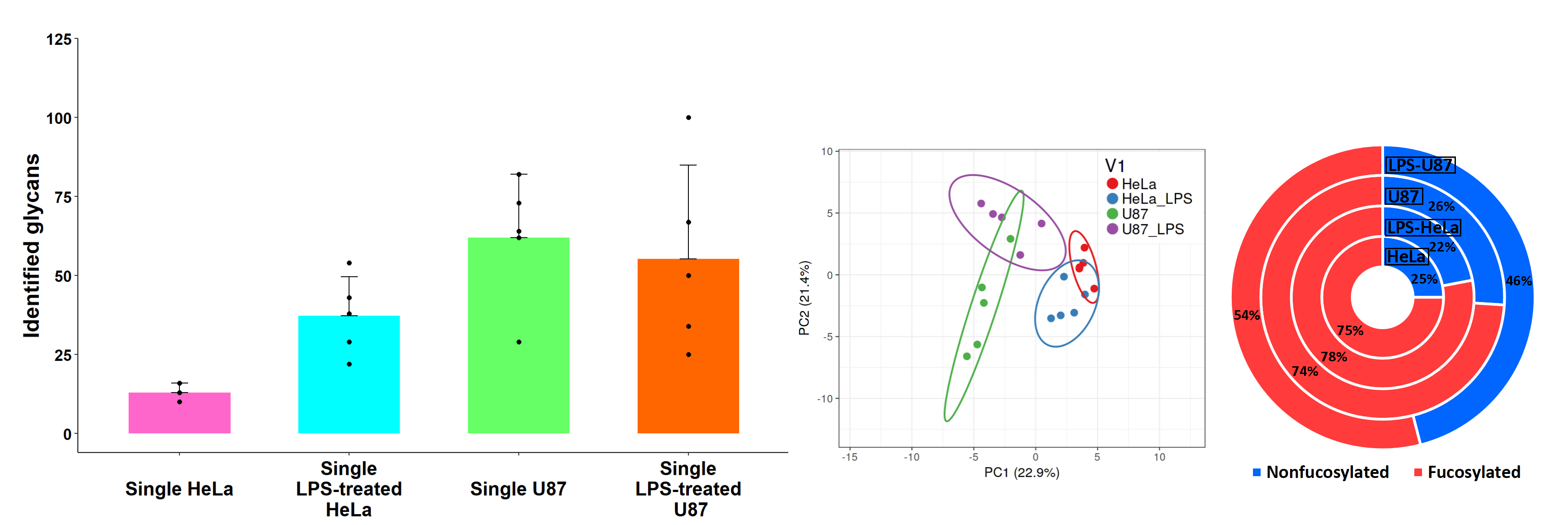
Fig. 2: CE-MS-based N-glycan profiling of single HeLa and U87 cells. Number of N-glycans detected and identified in single HeLa and U87 cells (left panel). Principal component analysis (PCA) of N-glycans detected in LPS-treated and untreated HeLa and U87 single cells showing a clear differentiation between both cell lines (middle panel). Fractional distributions of fucosylated N-glycans detected in HeLa and U87 single cells before and after LPS treatment demonstrating a downregulation of fucosylated N-glycans in LPS-treated U87 cells (right panel).
The developed workflow also resulted in the detection of over 170, 140, 370, and 220 N-glycans in isolates of total serum IgM, serum IgG, total plasma, and EVs, respectively, using ng-levels of serum proteins and nL/pL-levels of plasma isolates (Fig. 3). The numbers of N-glycans identified in the four types of analyzed blood-derived isolates shown here vastly exceed (~7-fold) those reported in other N-glycan profiling studies of similar complexity blood-derived isolates. A qualitative and quantitative comparative analysis of N-glycans detected in the four types of blood isolates was conducted with an exhaustive list of 679 glycans and demonstrated the uniqueness and high complexity of the four examined N-glycomes (Fig. 3).
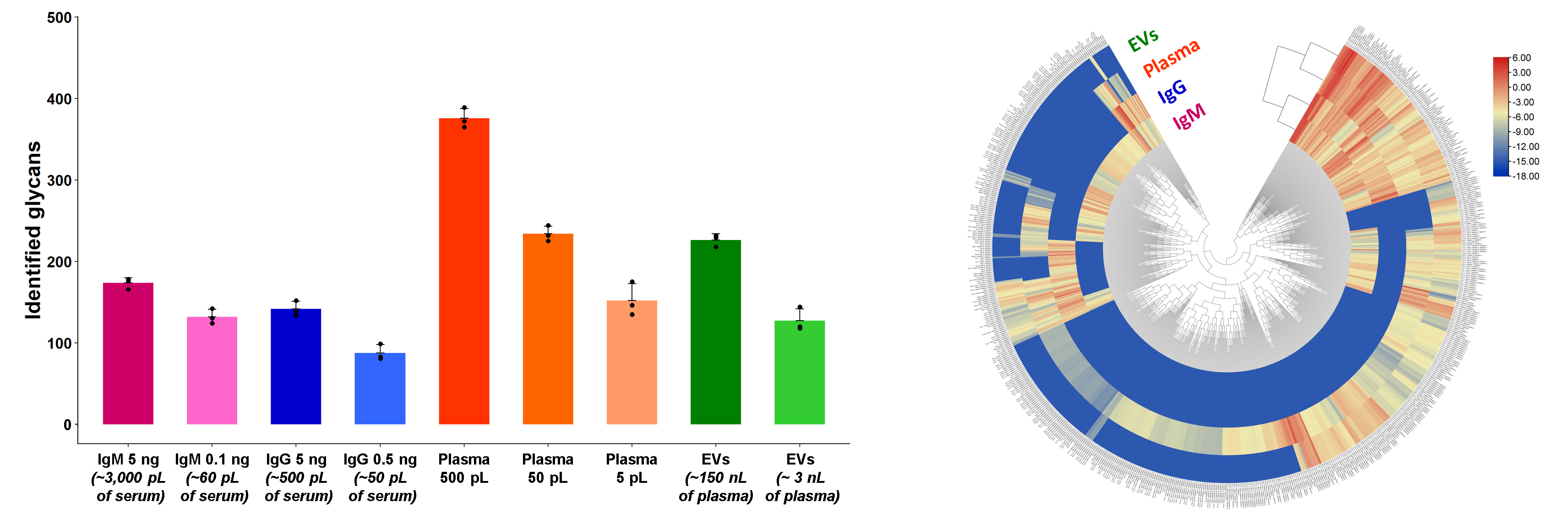
Fig. 3: CE-MS-based N-glycan profiling of human blood isolates. Number of N-glycans detected and identified in total serum IgM, serum IgG, total plasma, and EV isolates (left panel). Euclidean distance-based hierarchical clustering of quantitative glycomic profiles of IgM, IgG, total plasma, and EVs, using injected amounts of 5 ng of IgM, 5 ng of IgG, 500 pL of plasma, and 50 nL of EV isolate (corresponding to ~150 nL of plasma), respectively (right panel).
We envision that the developed high-sensitivity CE-MS-based workflow can open new doors in the field of glycomic profiling of scarce samples and single-cell glycomic research. For basic biology and clinical research, the technique can be used for cell phenotyping and studying the role of glycosylation in health and disease. In drug discovery and development, glycoprofiling technologies can monitor the drug glycosylation and be part of the quality control for new modalities of biotherapeutics, such as cell-based therapy. In glycomedicine, where therapeutics target glycans and glycosylation, glycomic profiling may assist in determining drug efficacy and specificity. In clinical applications, it can facilitate the detection of established disease biomarkers at early stages and the discovery of novel biomarkers for efficient and accurate disease diagnosis, prognosis, and treatment monitoring.
References:
1 Marie, A. L. et al. High-Sensitivity Glycan Profiling of Blood-Derived Immunoglobulin G, Plasma, and Extracellular Vesicle Isolates with Capillary Zone Electrophoresis-Mass Spectrometry. Anal Chem 93, 1991-2002, doi:10.1021/acs.analchem.0c03102 (2021).
2 Marie, A. L., Ray, S. & Ivanov, A. R. Highly-sensitive label-free deep profiling of N-glycans released from biomedically-relevant samples. Nat Commun 14, 1618, doi:10.1038/s41467-023-37365-4 (2023).
3 Johnson, K. R., Gao, Y., Gregus, M. & Ivanov, A. R. On-capillary Cell Lysis Enables Top-down Proteomic Analysis of Single Mammalian Cells by CE-MS/MS. Anal Chem 94, 14358-14367, doi:10.1021/acs.analchem.2c03045 (2022).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in