TrimNN: A New Way to Explore the Organization of Cells
Published in Protocols & Methods and Cell & Molecular Biology

Imagine walking into a bustling city. People aren’t just randomly scattered—they gather in coffee shops, work in offices, and cluster in parks. The way they group together says a lot about the city’s culture and health. Now, swap people for cells, and you get a sense of the challenge scientists face when studying how cells organize themselves inside tissues.
The layout of these “cellular neighborhoods” isn’t just for show—it influences how organs function, how diseases spread, and even how well treatments work. But until recently, we didn’t have a good way to capture the patterns in these microscopic communities. That’s where TrimNN comes in.
Why Cell Arrangement Matters
Cells in our body aren’t loners. Immune cells talk to each other to coordinate defenses. Neurons form intricate networks to process information. Cancer cells interact with their surroundings in ways that help them grow or evade the immune system. Understanding these spatial arrangements could reveal early signs of disease or help design more targeted therapies.
In the past, scientists often used a “top-down” approach: group cells into broad categories and look for general patterns in how those categories appear together. It’s a bit like describing a city by saying, “20% of the people here work in finance, 30% in hospitality,” without noting where those people meet or how they interact.
This works up to a point, but it misses the fine details—the equivalent of knowing that the city’s best ideas might be born in the corner coffee shop where coders and designers regularly meet.
From Big Groups to Building Blocks
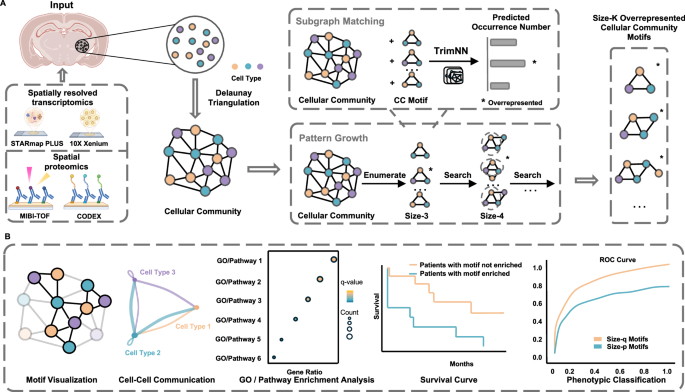
TrimNN flips the script. Instead of starting with large groupings, it takes a bottom-up approach, looking for tiny recurring patterns of cells, called cellular community (CC) motifs.
A CC motif is like a “mini-neighborhood” of cells—a handful of cells of specific types, arranged in a particular way. Some motifs might be as small as a single cell type; others are small constellations of different cell types linked together. These motifs are the building blocks of tissue organization.
Why focus on these small patterns? Because they can be conserved—recurring again and again in healthy tissues—or they can shift in diseases, revealing clues about what’s going wrong.
The Challenge: Finding Patterns in a Sea of Data
Finding these motifs is no small feat. Modern spatial omics technologies can measure gene or protein activity in thousands of cells and record their exact positions. That’s like having a map of every building in the city along with the job title of every person inside.
The problem? Sifting through all the possible patterns of connections between cell types is computationally enormous. Traditional “subgraph matching” methods (think: pattern-finding in networks) slow to a crawl with anything more than a few cell types. This is why most previous studies only looked at the tiniest motifs—like pairs or triplets of cells.
Introduce TrimNN
TrimNN (Triangulation cellular community motif neural network) is a graph-based deep learning framework that address this challenge.
It works by:
-
Turning cells into a network
Each cell is a “node,” connected to its nearest neighbors based on physical closeness. This network is shaped using a technique called Delaunay triangulation, which connects cells into triangles—ideal for capturing local relationships. -
Breaking the big problem into smaller ones
Instead of trying to count all motifs at once (which is painfully slow), TrimNN breaks the job into many smaller “yes/no” questions: Does this small pattern appear in this part of the network? -
Using smart pattern recognition
It employs a type of graph neural network (GIN) combined with “positional encoding” to understand not just who the neighbors are but also how far apart they are. -
Growing motifs step-by-step
Once TrimNN finds smaller motifs, it can “grow” them into bigger ones by adding more cells—letting scientists explore more complex arrangements without drowning in computation.
The result? TrimNN can spot large, meaningful motifs in seconds—something older methods couldn’t realistically do.
What We Learned from TrimNN
The team tested TrimNN on both simulated datasets and real-world biological data. Here are some of the highlights:
-
Predicting Cancer Outcomes
In a study of colorectal cancer patients, TrimNN found specific motifs—combinations of macrophages (a type of immune cell) and smooth muscle cells—whose arrangement could predict patient survival. Interestingly, simply knowing how many of each cell type was present wasn’t enough; it was their spatial arrangement that made the difference.
Some larger motifs, like two macrophages next to two smooth muscle cells, were strongly linked to worse survival. This suggests that how these cells “cluster” could influence how tumors progress.
-
Shedding Light on Alzheimer’s Disease
In Alzheimer’s mouse brains, TrimNN revealed two distinct kinds of motifs involving microglia (immune cells in the brain) and excitatory neurons. Some motifs tended to appear away from amyloid-beta plaques, while others were tightly co-located with them. These differences matched known biology—microglia often gather near plaques and can either help clean them up or worsen damage.
TrimNN also showed that these motifs had different communication “signatures” between cells, and distinct sets of active genes, potentially revealing different roles in disease progression.
-
Understanding Tumor-Immune Interactions
In colorectal carcinoma, TrimNN identified what the researchers called Shifted Interaction Motifs—patterns that change dramatically from healthy to cancerous tissue as more cell types join the group. These shifts may reflect how the tumor rewires its microenvironment to evade immune attacks.
Why This Matters
By breaking tissue organization into these repeatable, interpretable motifs, TrimNN offers:
- Better disease markers: Motifs can serve as “fingerprints” of disease state, more robust than cell counts alone.
- Biological insight: Because motifs are small and interpretable, researchers can connect them to known cell behaviors, pathways, or gene activity.
- Cross-dataset generalizability: Unlike some machine learning models that depend heavily on the training data, motifs are concrete features that can be compared across studies.
A Tool for the Spatial Biology Era
Spatial biology is one of the fastest-growing areas in biomedical research. As datasets get bigger and more complex, tools like TrimNN will be essential for turning raw maps of cells into actionable biological knowledge.
In many ways, CC motifs are to tissues what words are to language: small, meaningful units that can be combined into bigger stories. By decoding this cellular “grammar,” we can start to read the narratives of health, disease, and treatment response written in the spatial fabric of our bodies.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in