Tumor microenvironment composition and developmental stemness features predict survival in pancreatic cancer
Published in Cancer

PDAC has remained largely refractory to available therapeutics. Numerous cell states are known to comprise the PDAC TME, however, the developmental stemness and co-occurrence of these cell states remains poorly defined. Over the past decade, bulk tumor sequencing has enabled annotation of the genomic landscape in PDAC and led to several classification systems. These systems consistently demonstrate the existence of two major subtypes of PDAC: the classical/pancreatic progenitor subtype associated with an improved survival, and the more aggressive basal subtype associated with a poorer prognosis. However, the existence of these subtypes alone has not translated to effective clinical interventions. In part due to the reliance of these methods on bulk sequencing data - creating blind spots in cell states and features of individual cells within a single tumor sample.
In this work, we take a closer look at the interplay of these subtypes with the TME in an effort to better personalize care and significantly improve outcomes for select PDAC patients. To do so, we performed scRNA-seq of PDAC from standard-of-care time-of diagnosis endoscopic ultrasound-guided fine needle biopsy (EUSFNB) specimens at the time-of-diagnosis (n=25) and from surgical samples (n=6) obtained from tumor resections.
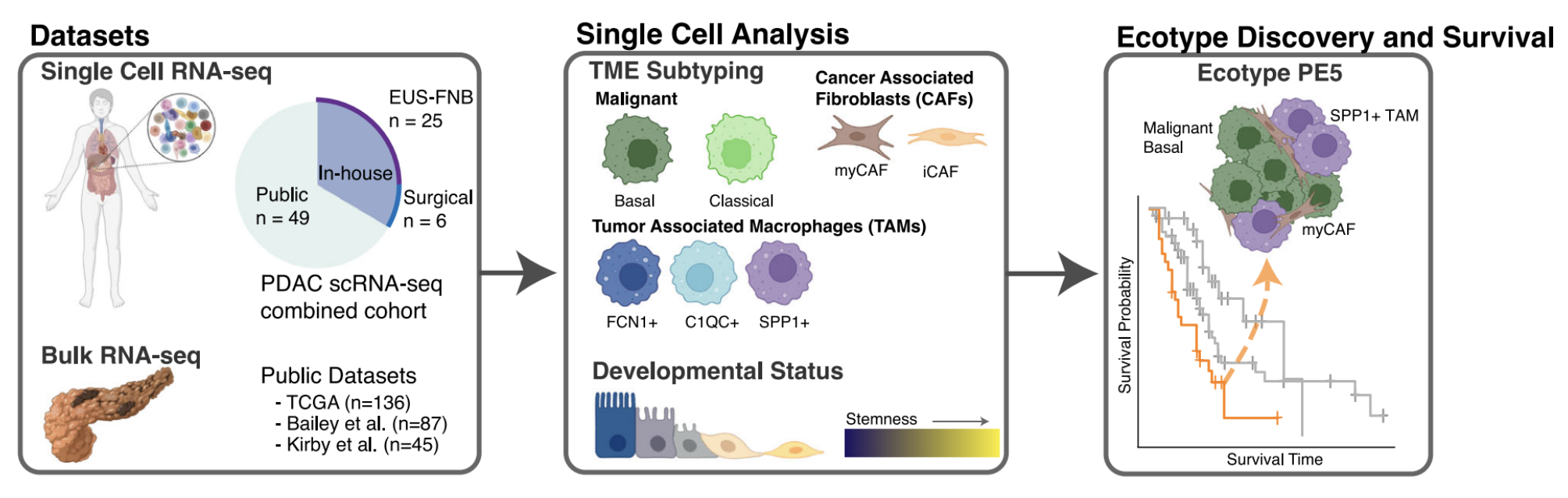
(Of note: in our experience scRNA-seq and downstream analyses were able to be performed on time-of-diagnosis EUSFNB samples within 2 weeks, which is generally in the time frame that patients are seen by their medical oncologists and treatment decisions are made.)
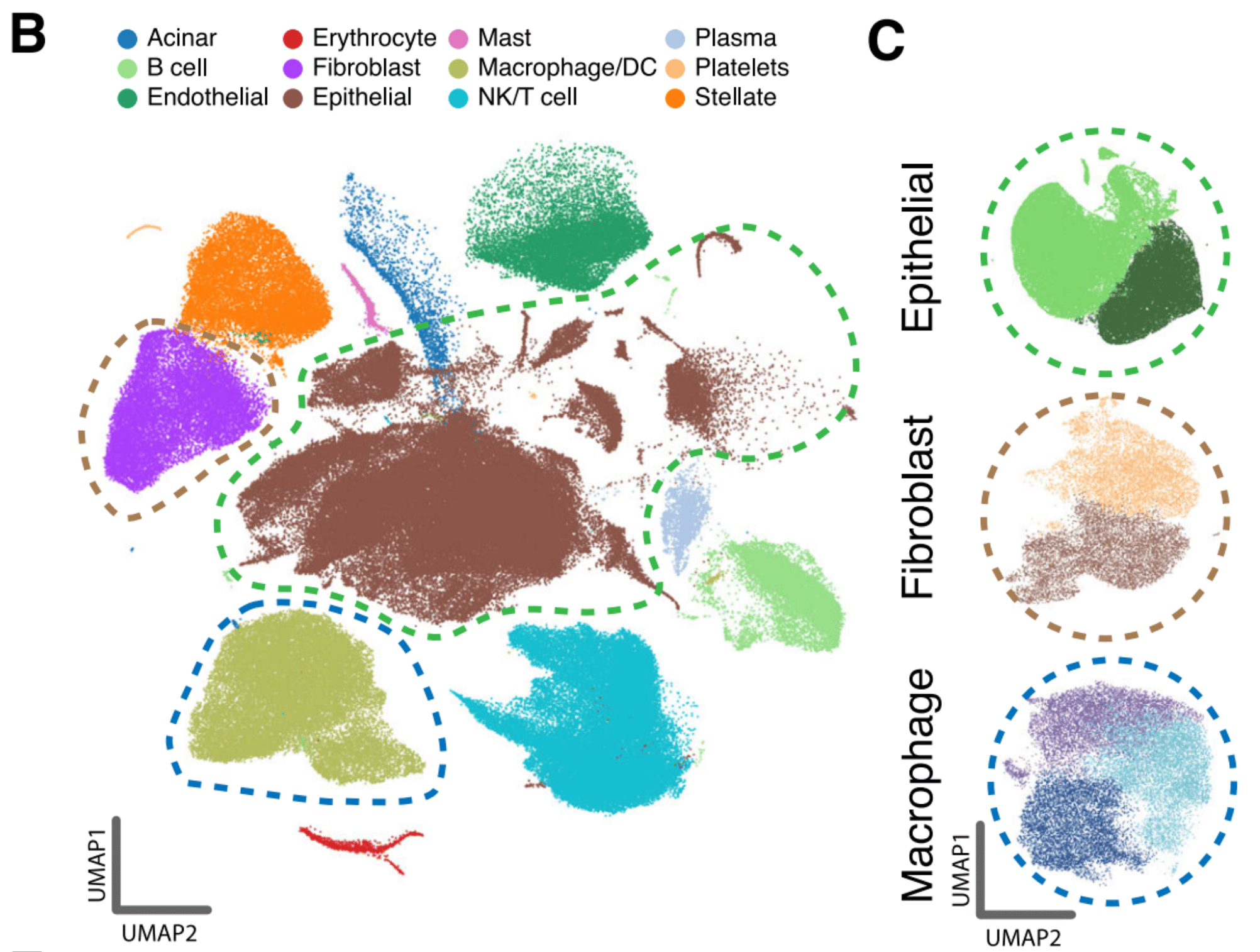
In total, we acquired 31,215 cells across 25 independent PDAC patients for our in-house EUS-FNB cohort and 11,353 cells from 6 independent PDAC patients for our in-house surgical cohort. To increase power, we then combined the in-house scRNA-seq data with three publicly available datasets (Peng et al., Chan-Seng-Yue et al., and Lin et al.) increasing our sample size to a total of 198k cells from 80 independent PDAC tumors.
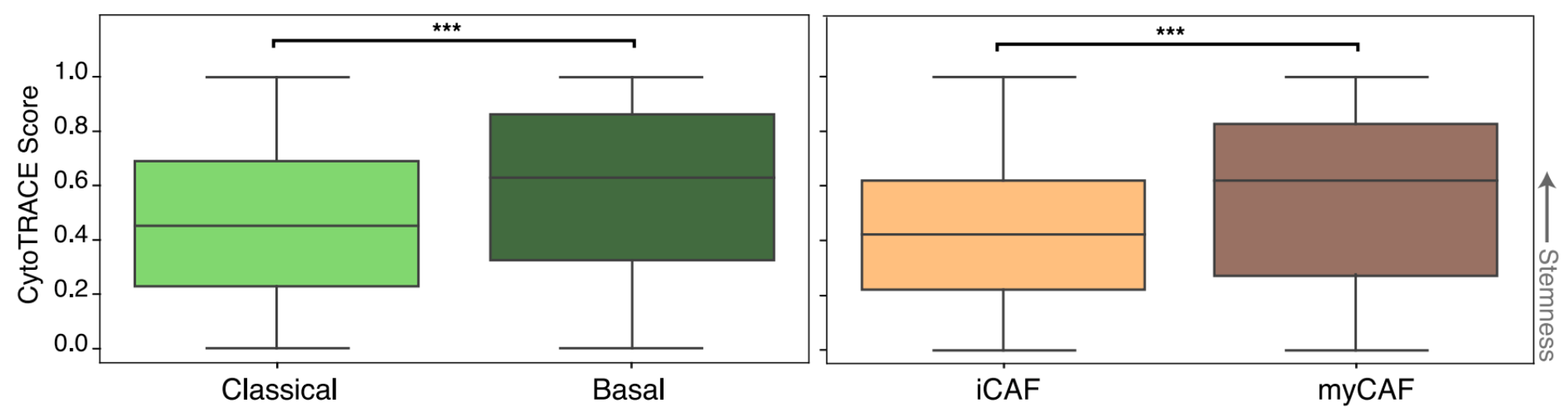
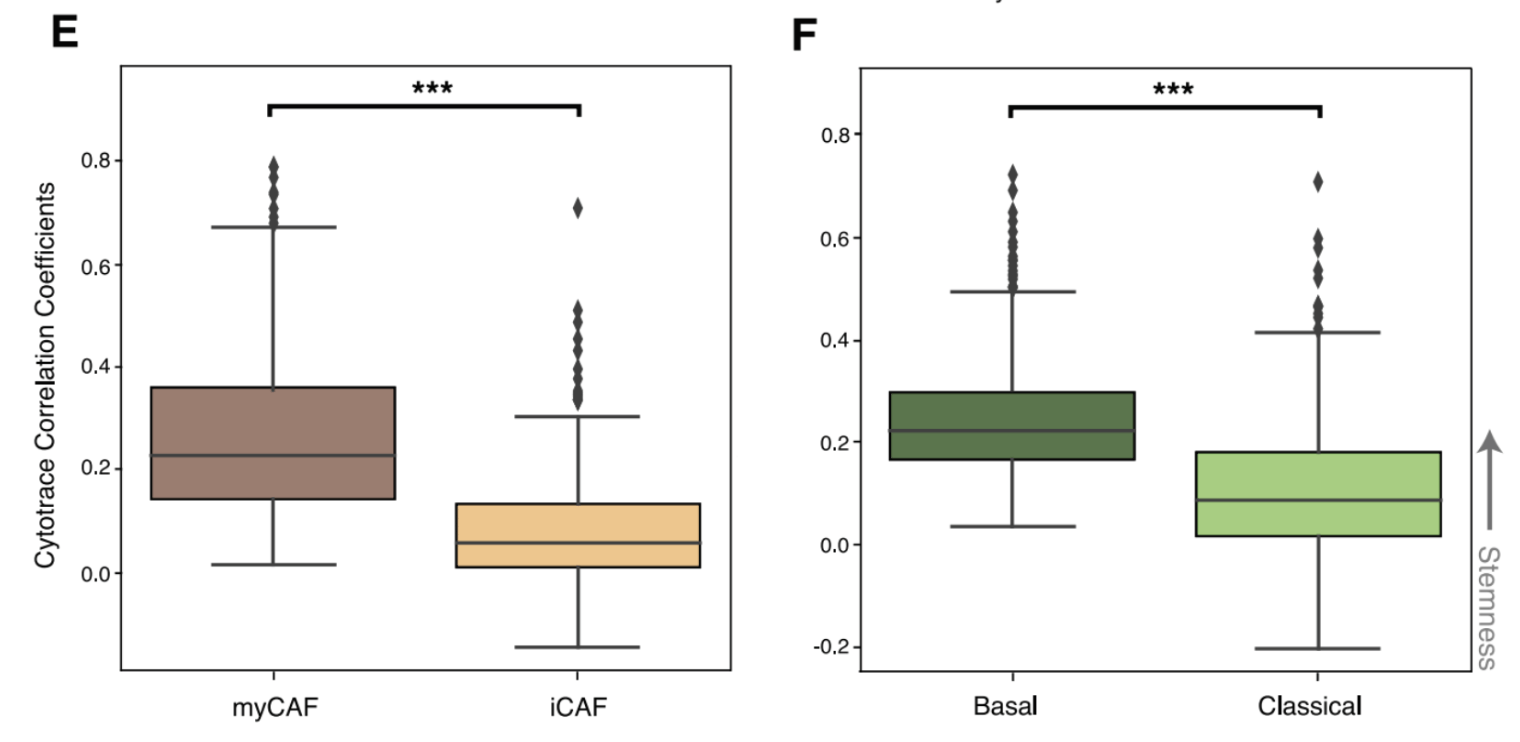
After identifying cell states in the single cell data, we then determined the developmental stemness of the malignant and CAF cell states using CytoTRACE (Gulati et al.). Basal-like malignant cells were more stem-like than their classical counterparts. This difference has been previously suggested in the literature, with the more aggressive basal-like subtype being more likely to undergo EMT, resulting in higher rates of metastasis. Interestingly, we also found that myCAFs are significantly more stem-like than iCAFs. (More on that later).
We then extended our single-cell expression profiles to publicly available bulk expression datasets (TCGA, Bailey et al., Kirby et al.) with associated clinical metadata to find cell state patterns associated with patient survival. For this, we modified the insilico TME dissection tool EcoTyper (Luca et al.) to allow for the specification of exactly predefined cell states. (In the published Ecotyper tool, cell states must be discovered de novo, meaning specific cell states cannot be defined upfront). We discovered three main pancreatic ecotypes, termed PE1, PE5, and PE6.
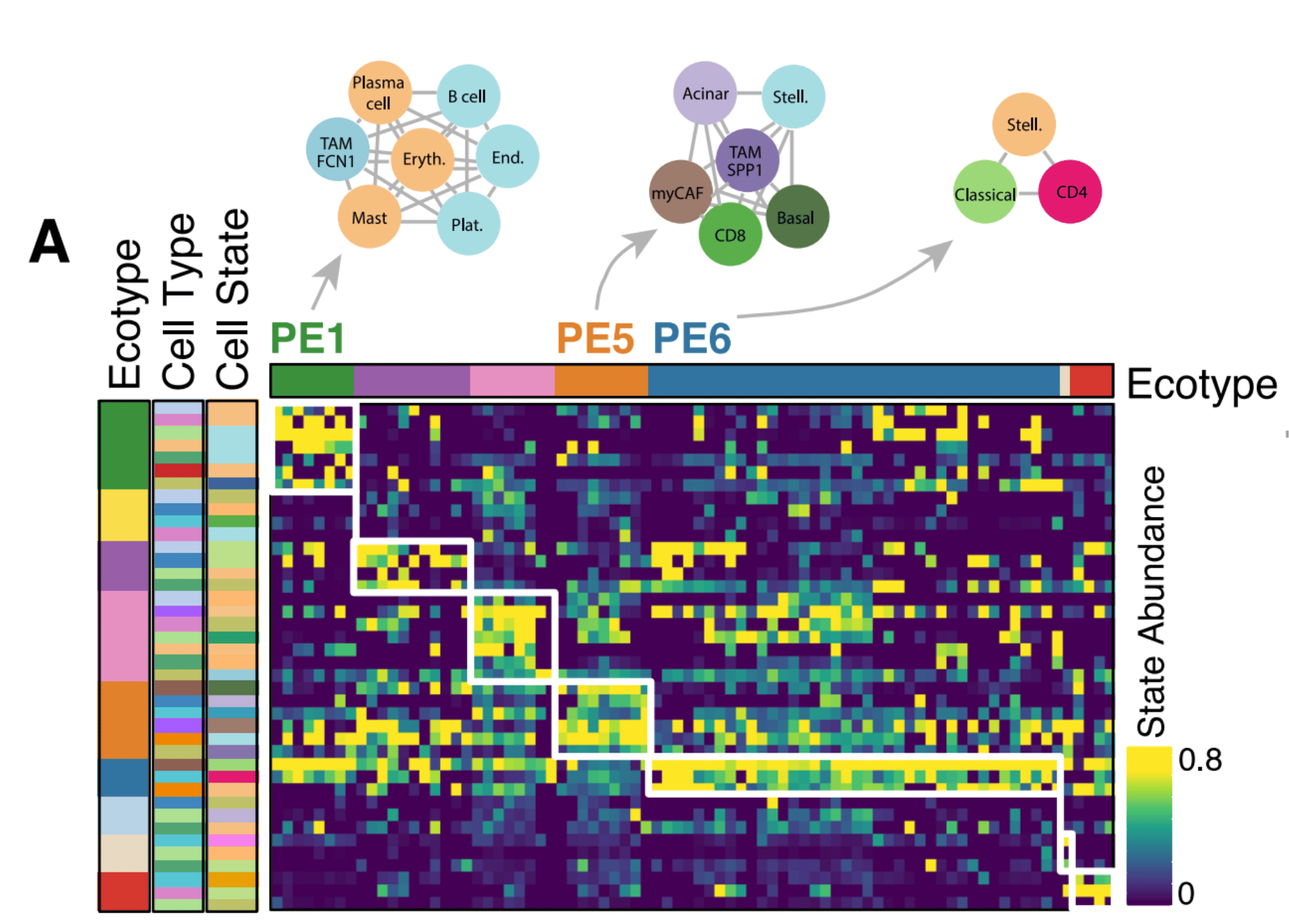
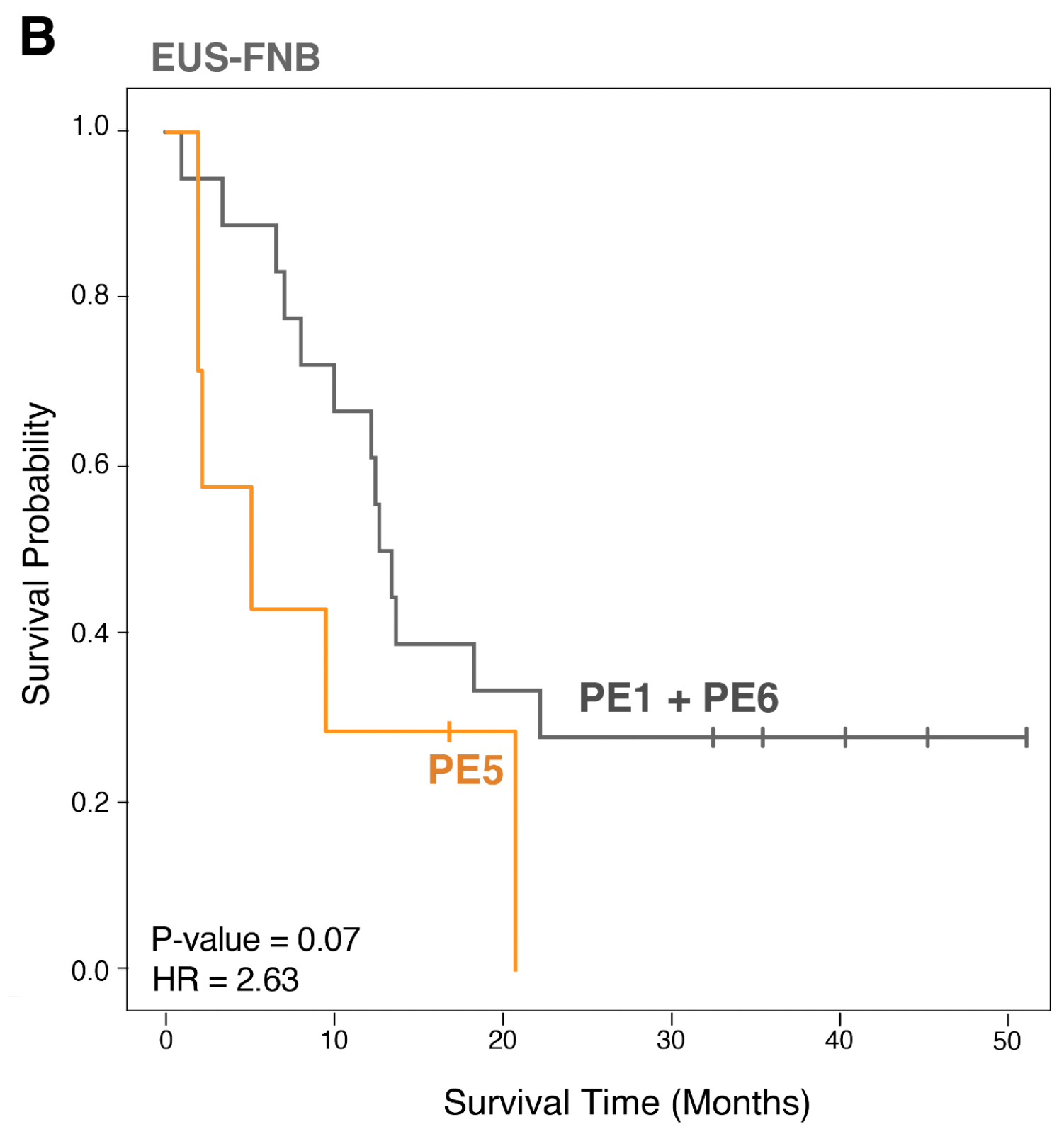
Notably, samples with a PE5-dominant ecotype showed consistently poor survival across all bulk RNA-seq datasets including at the time of diagnosis in the EUS-FNB cohort. PE5 was additionally enrichment for cell states known to be associated with aggressive tumor behavior, including basal-like tumor cells, myCAFs, and SPP1+ TAMs.
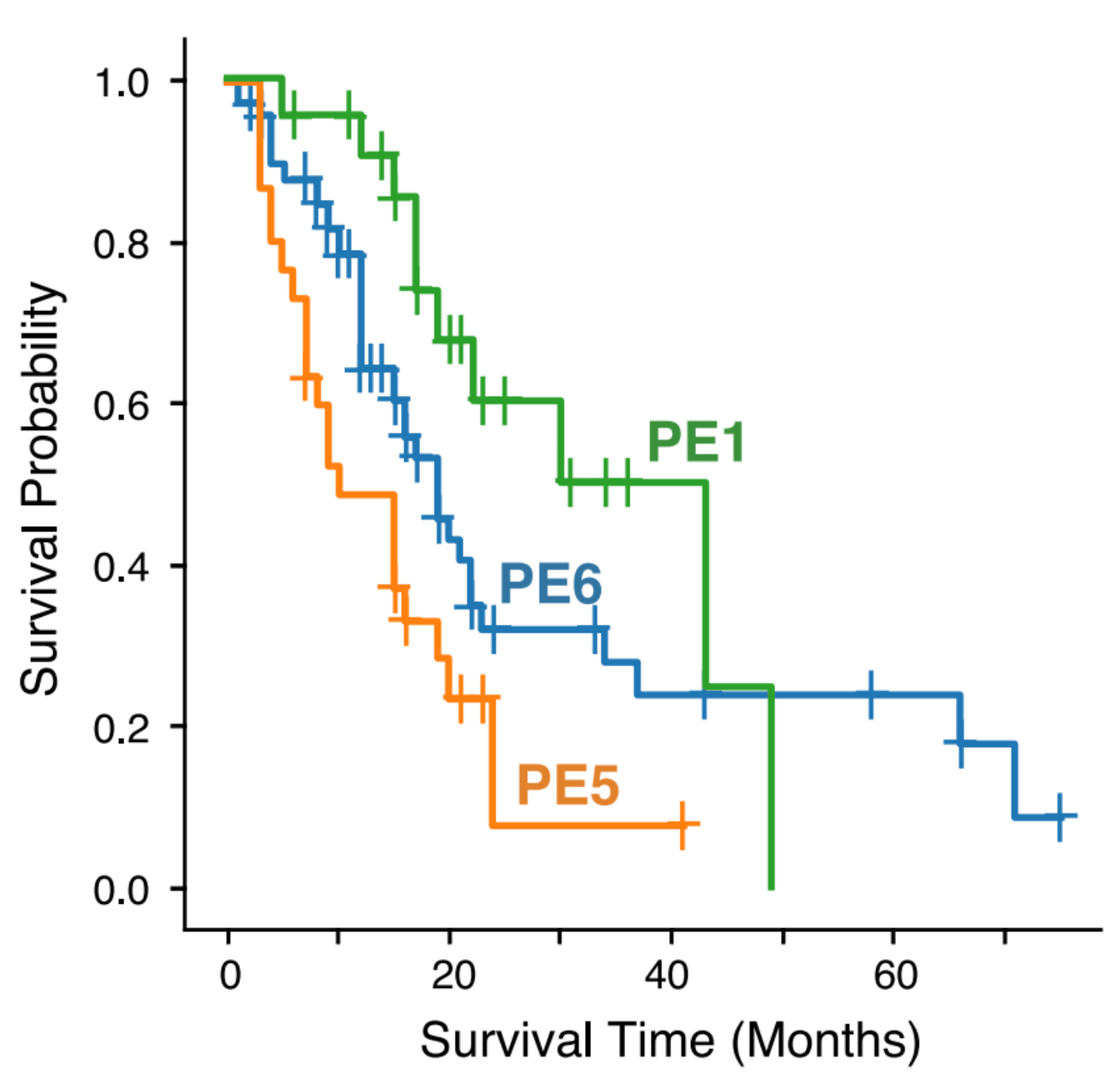
In contrast, PE1 and PE6 were associated with better survival outcomes than PE5. PE1 is immune enriched, containing plasma cells, mast cells, B cells, and FCN1+ TAMs, while PE6 contains CD4 T cells and classical malignant cells.
Importantly, PE5’s negative association with survival was also present in EUS-FNB samples taken at the time of diagnosis, opening the possibility of more upfront clinical decision-making in the future.
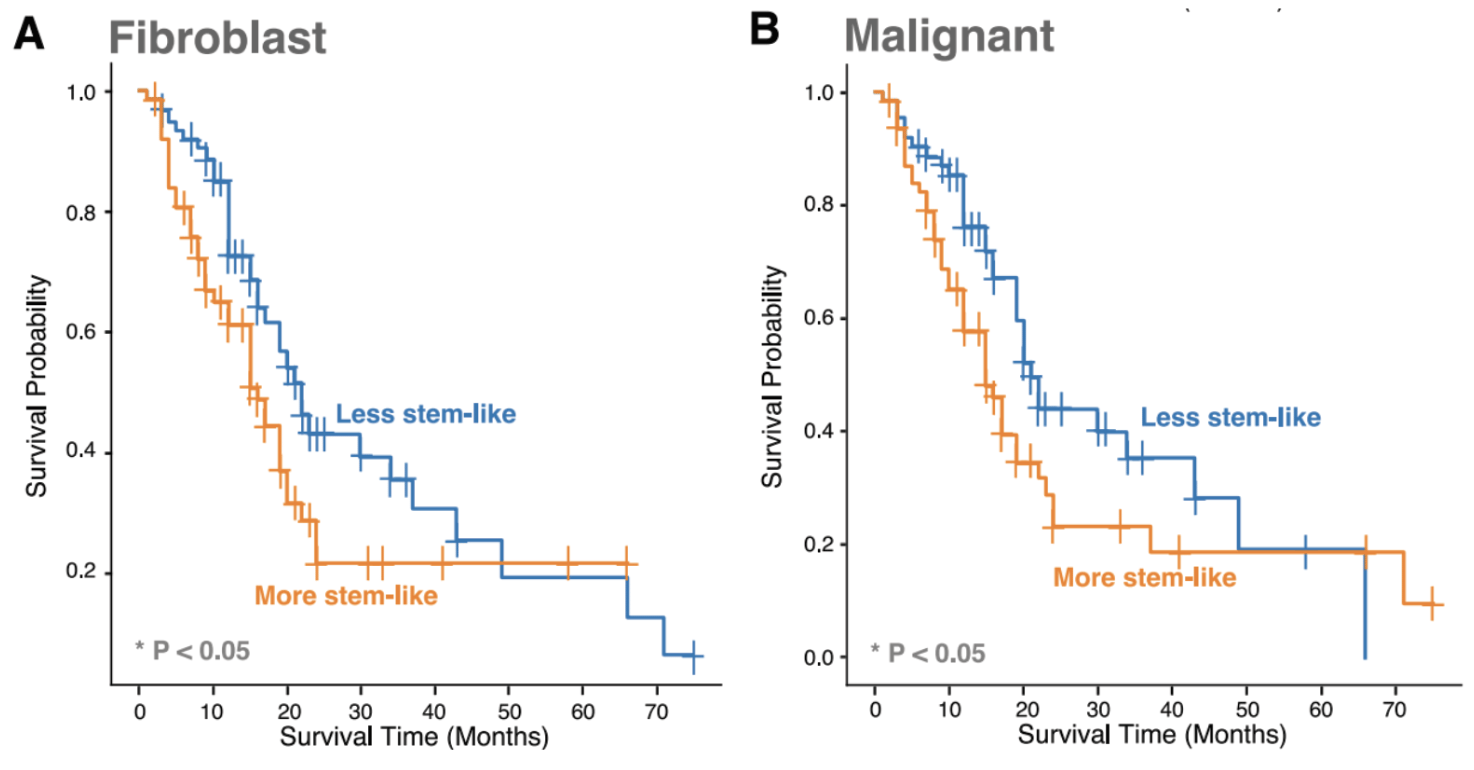
Given the developmental continuum in CAF and malignant cell states found in our single-cell data, along with pancreatic ecotype PE5’s enrichment for myCAFs and basal-like malignant cells, we more directly quantified the impact of stemness on patient survival for these cell states. To do so, we calculated a developmental stemness score for bulk RNA-seq samples. When partitioned into low vs. high stemness groups based on this score, we observed poor survival with more stem-like CAFs and malignant cells.
Notably, the CAF and malignant states associated with PE5 (myCAF and malignant basal-like) were significantly more enriched for stemness correlated genes than non-PE5 states.
In summary, we identified pancreatic ecotypes and developmental continuums from PDAC RNA sequencing data, including time-of-diagnosis EUS-FNB specimens, that revealed connections between TME composition, stemness, and patient survival that could lead to better upfront risk stratification and more personalized clinical decision making in the future.
Follow the Topic
-
npj Precision Oncology

An international, peer-reviewed journal committed to publishing cutting-edge scientific research in all aspects of precision oncology from basic science to translational applications to clinical medicine.
Related Collections
With Collections, you can get published faster and increase your visibility.
AI Approaches in Drug Design
Publishing Model: Open Access
Deadline: Mar 31, 2026
Genomic Instability
Publishing Model: Open Access
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in