Uncovering the origins of H58 to stop drug-resistant typhoid in its tracks
Published in Microbiology
Since the 1940s, antibiotics have been used to treat typhoid. With the rise of drug-resistant Salmonella Typhi (S. Typhi), the bacteria that causes typhoid, current antibiotics may not effectively treat typhoid for much longer. Indeed, resistance to every antibiotic used to treat S. Typhi has been reported in South Asia, a dire trend that will likely have deadly consequences. To address this growing threat more effectively, we need to understand where and how drug resistance starts. The growing focus on genomic surveillance monitors the emergence and spread of drug-resistant typhoid.
What is genomic surveillance?
Genomics is the study of the structure, function, evolution, and mapping of genomes – a set of genes or genetic material present in a cell or organism. Genomics has been widely used to track infectious diseases through characterization of pathogen’s genetic material. Genomic surveillance is the process of continuously monitoring pathogens and analyzing their genetic similarities and differences to make interpretations about relatedness and spread, as well as their drug resistance profiles.
Where did H58 come from?
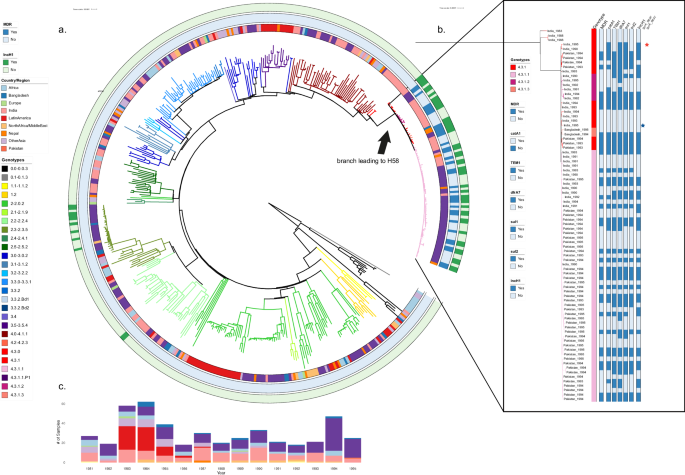
Genomic analysis helped to demonstrate the global spread of a specific lineage of multi-drug resistant (MDR) typhoid called Haplotype 58 or H58, the lineage from which extensively drug resistant (XDR) S. Typhi emerged. Researchers set out to analyze whole genome sequences of S. Typhi isolated from travelers returning to the United Kingdom, starting from the estimated time of H58 emergence, to better understand when and where this globally important lineage originated.
Our analysis includes the earliest publicly available H58 S. Typhi genomes and suggests that these organisms, which were originally MDR, emerged in India in 1987 before spreading further through South Asia and then globally. During subsequent decades, H58 rapidly established a global presence, radiating outwards from South Asia to other regions. It is the only major S. Typhi lineage to have spread across borders so quickly.
These early organisms were very genetically distinct from their ancestral organisms, possessed mutations that allowed them to survive and become more virulent in the presence of bile. These and other genetic characteristics suggest that the first H58 organism may have been generated during chronic carriage in the gallbladder. The potential for hypermutation and shedding of S. Typhi from chronic carriers could result in the emergence of additional S. Typhi organisms with the potential for global spread.
The apparent ability of H58 organisms to rapidly acquire and maintain drug resistant genes poses a monumental threat to global health. The earliest H58 organisms already exhibited resistance to first-line antibiotics, and soon mutations associated with decreased susceptibility to ciprofloxacin, a commonly prescribed antibiotic, appeared within this linage. Researchers have reported H58 organisms with resistance to all oral antibiotics used to treat typhoid fever.
Genomic data continue to show that drug-resistant typhoid most frequently emerges in and spreads from South Asia, a region where population density, a lack of access to clean water and improved sanitation, and the effects of climate change may result in even higher risk of typhoid transmission. Prioritizing immunization in South Asia may not only reduce transmission but also slow the emergence and spread of drug resistant typhoid.
What can we do about it?
TCVs represent a crucial tool to combat drug-resistant typhoid. By protecting individuals from infection, TCVs can reduce the incidence of disease caused by drug-resistant S. Typhi. If an individual is protected by TCV and avoids a course of antibiotic treatment, this reduces pathogen exposure to antibiotics, lowering the chances of the development of resistance. Thus, through prevention of infection and potential reduction of antibiotic use, vaccines can limit the emergence and spread of drug-resistance. In addition, screening for and treating chronic carriers may result in lower risk of additional emergence of drug-resistant typhoid.
Addressing rapidly rising prevalence and severity of drug resistance is an urgent global health priority. Through collaborative drug resistance surveillance efforts such as the Global Typhoid Genomics Consortium, we can inform updated typhoid treatment guidelines and the prioritization of preventative interventions like TCVs. Ongoing genomic sequencing furthers our understanding of how drug-resistant S. Typhi spreads so policymakers can make informed decisions about typhoid prevention and contro
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in