Unleashing the Potential of CRISPR for In Vivo Cancer Research
Published in Cancer, Microbiology, and Protocols & Methods

CRISPR technology has taken cancer research by storm, especially in generating targeted mutations in cell lines. We utilized CRISPR in vivo to investigate lung, brain, and prostate cancer by delivering CRISPR guides with viral particles to Cas9 transgenic mice1-4.
In this study, we took a bold approach, simultaneously mutating eight genes related to human prostate cancer 5. This resulted in the development of complex tumors with multiple mutations, revealing positive genetic interactions in cancer development. Strikingly, cancer development was very rapid, and mice reached humane endpoints 8 weeks after tumor initiation, due to a large primary tumor. Delineation of driver mutations revealed that the loss of the less common tumor suppressor genes Stk11 and RNaseL accelerated cancer progression in combination with the loss of Pten, Rb1, and Trp53. Next, we added three sgRNAs targeting epigenetic factors along with the aforementioned five guides, targeting in total 8 genes. Introduction of additional mutations in 3 epigenetic factors did not alter tumor progression even though all three genes are often found mutated in prostate cancer. Instead, metastasis formed in the lungs of all the mice, which was not observed when only the five tumor suppressor genes were mutated. This strongly indicated that epigenetic modification could drive dissemination of primary tumor cells and establishment of a secondary tumor.
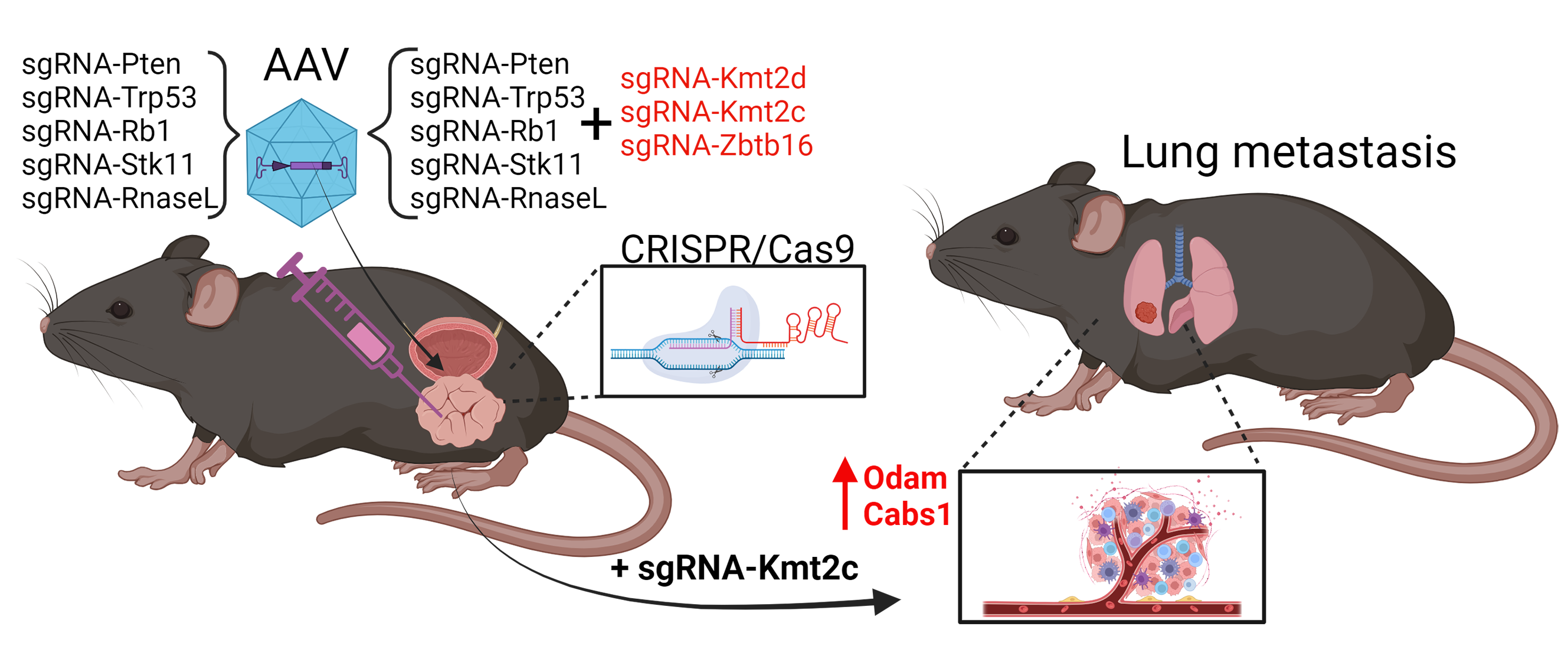
To delineate the molecular mechanisms behind metastasis formation, different approaches were employed. First, we performed whole-genome sequencing to evaluate if new somatic mutations had occurred in the metastasis. Although we found approximately 1.5 mutations per million bases, only few were present in coding sequences and fewer were shared between more than one sample. Therefore, we performed RNA sequencing to examine changes in gene expression. In general, the expression in the tumor samples differed tremendously from normal prostate tissues. Surprisingly, fewer than 30 genes were differently expressed between tumors with mutated epigenetic factors and those with intact ones. Interestingly, a conserved cluster of five genes was strongly upregulated by the loss of Kmt2c, which we validated in a human prostate cell line. None of the five upregulated genes had previously been associated with cancer, but we mutated two of the genes, Odam and Cabs1, in a mouse prostate cell line and orthotopically implanted them in mice. The two mutated genes impaired metastasis formation and growth. We did not unveil the molecular alterations of Odam and Cabs1, but our work shows that the loss of Kmt2c facilitates metastasis formation, partly by upregulating a conserved gene cluster including Odam and Cabs1.
Research carried out in mice can be challenging to translate to humans. However, as we studied genes commonly mutated in human prostate cancer and validated the findings in human prostate cell lines, this suggests that the results can be applied to human cancer. To gain further insight into human relevance, we investigated dysregulated genes between primary and secondary tumors in mice. Hereby, we identified several sets of genes that could differentiate human primary and secondary prostate cancer.
Altogether, by using CRISPR technology in vivo, we generated a mouse model of prostate cancer where we could unveil the function of numerous commonly mutated genes. Furthermore, this analysis also differentiated genes that promote tumor growth and genes that regulate cell fate and dissemination. Hopefully, this will pave the way for future studies based on CRISPR in vivo editing to reveal the function of less explored genes in cancer and help gain a better understanding of human cancer.
1 Joseph, J. V. et al. STING activation counters glioblastoma by vascular alteration and immune surveillance. Cancer Lett 579, 216480 (2023). https://doi.org:10.1016/j.canlet.2023.216480
2 Cai, H. et al. In Vivo Application of CRISPR/Cas9 Revealed Implication of Foxa1 and Foxp1 in Prostate Cancer Proliferation and Epithelial Plasticity. Cancers 14, 4381 (2022). https://doi.org/10.3390/cancers14184381
3 Riedel, M. et al. In vivo CRISPR inactivation of Fos promotes prostate cancer progression by altering the associated AP-1 subunit Jun. Oncogene 40, 2437-2447 (2021). https://doi.org:10.1038/s41388-021-01724-6
4 Berthelsen, M. F. et al. Comparative Analysis of Stk11/Lkb1 versus Pten Deficiency in Lung Adenocarcinoma Induced by CRISPR/Cas9. Cancers (Basel) 13 (2021). https://doi.org:10.3390/cancers13050974
5 Cai, H. et al. CRISPR/Cas9 model of prostate cancer identifies Kmt2c deficiency as a metastatic driver by Odam/Cabs1 gene cluster expression. Nat Commun 15, 2088 (2024). https://doi.org:10.1038/s41467-024-46370-0
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Ask the Editor – Inflammation, Metastasis, Cancer Microenvironment and Tumour Immunology
Got a question for the editor about inflammation, metastasis, or tumour immunology? Ask it here!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in