Unlocking the potential of AI for studying leaf stomata: A valuable image dataset for ecologists and plant scientists
Published in Plant Science and Anatomy & Physiology

The need for a comprehensive stomatal image dataset
Studying stomatal responses to environmental factors, such as humidity and soil moisture, is crucial for understanding plant behavior, ecosystem dynamics, and even climate effects. Traditional methods of manually counting and measuring stomatal properties have limitations in terms of dataset size and scalability.
To overcome these limitations, there's a growing interest in leveraging AI, particularly deep learning and convolutional neural networks (CNNs), for stomatal detection and measurement. One of the noteworthy CNN architectures used for this purpose is "You Only Look Once" (YOLO). However, the effectiveness of AI models heavily relies on the quality and quantity of training data.
Introducing the stomatal image dataset
Researchers from the Department of Forestry at Mississippi State University and the School of Geography at Nanjing Normal University have compiled a valuable stomatal image dataset. This dataset includes around 11,000 unique images collected from temperate broadleaf angiosperm trees, specifically from 17 hardwood species and seven Populus taxa.
For each image in the dataset, the researchers meticulously labeled two important stomatal components: inner guard cell walls and the entire stomata (stomatal aperture and guard cells). To make this data accessible to the scientific community, YOLO label files were created for each image.
Benefits of the dataset
This dataset offers several key advantages for researchers, ecologists, and plant biologists: High-Quality Training Data: With around 11,000 labeled images, the dataset provides a robust foundation for training and optimizing machine learning models, such as YOLO, to detect, count, and measure stomata accurately.
Diversity of Stomatal Characteristics: Researchers can explore the wide range of stomatal characteristics across different hardwood tree species, enhancing our understanding of how environmental factors affect stomatal behavior.
Innovative Stomatal Research: The dataset opens the door to developing novel indices for measuring stomatal properties, potentially leading to breakthroughs in ecological and plant science.
Data collection and validation
The researchers collected stomatal images from various hardwood species and Populus taxa. Each leaf was carefully prepared for stomatal observation, and images were captured using advanced microscopy techniques. The labeling process involved both manual and pre-trained model labeling methods, ensuring the accuracy of the annotations.
To validate the quality of the dataset, the researchers used YOLOv7 and YOLOv8 models. The results demonstrated high precision and recall values, confirming the dataset's suitability for machine learning model training.
Accessing the dataset
The stomatal image dataset is publicly available on figshare (https://doi.org/10.6084/m9.figshare.22255873) and Zenodo (https://doi.org/10.5281/zenodo.8266240), making it easily accessible to researchers worldwide. The dataset comprises original images, labels, and essential data records. These records include critical information such as species names, magnification, image dimensions, and resolution, all of which are essential for studying stomatal characteristics.
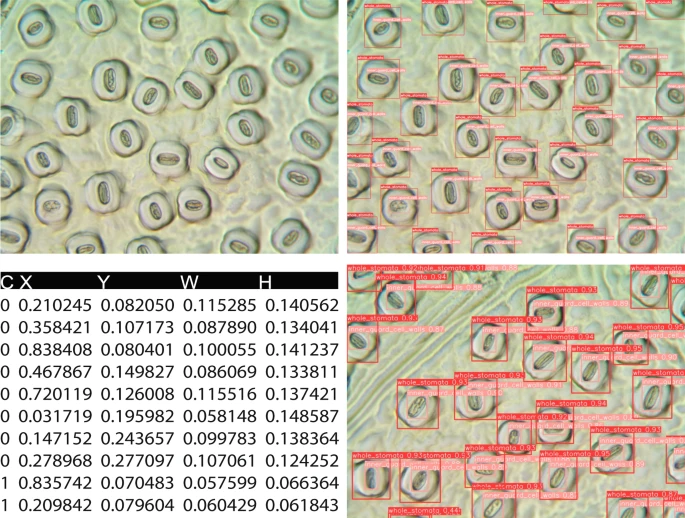
Fig. 1. Original and annotated leaf stomatal images and the label file structure. C, X, Y, W, H represent class, x_center, y_center, width, and height of the bounding boxes, respectively. The x_center and y_center are expressed as normalized coordinates that correspond to the center of the bounding box, while width and height are normalized values that represent the relative width and height of the box concerning the dimensions of the image. Note that "C, X, Y, W, H" do not exist in label files, and we used these headings for explanation.
Using the dataset
To harness the full potential of this dataset, researchers are encouraged to upload the images and labels to platforms like Roboflow. This allows for annotation verification and format conversion, ensuring the dataset is tailored to individual research needs.
By using AI models trained on this dataset, researchers can extract valuable information about stomatal properties, enabling the development of new indices for stomatal assessment. For example, measurements like stomatal area and guard cell dimensions can be estimated from the bounding boxes identified by the YOLO model. For example, using this dataset, Jiaxin Wang et al. have developed StoManager1 (available on: https://github.com/JiaxinWang123/StoManager1), which can be used as a high through-put automated labeling and measuring tools for stomatal and guard cell metrics.
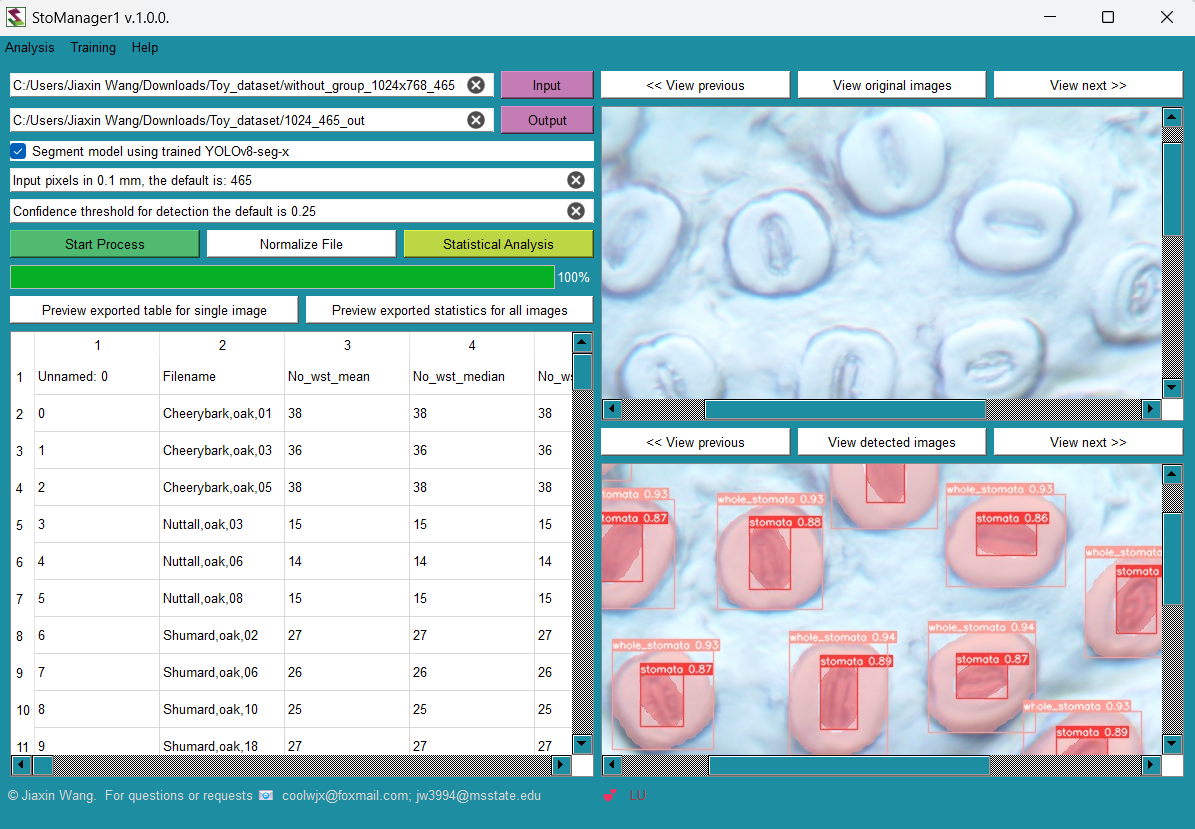
Fig. 2. User-friendly interface of StoManager1.
In conclusion, this comprehensive stomatal image dataset opens up exciting opportunities for advancing research in ecology, plant biology, and ecophysiology. By leveraging the power of AI, researchers can unlock new insights into the behavior of stomata, ultimately contributing to our understanding of plant responses to the environment and their impact on ecosystems and climate.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in