Unraveling the Causes of Multiple Early-Stage EGFR-Mutant Lung Cancers
Published in Cancer and Genetics & Genomics

Motivation
Lung cancer is a major killer, responsible for about 12% of all cancer diagnoses and 1.8 million deaths each year worldwide1. The vast majority of these cases are non-small cell lung cancer (NSCLC)2.
When NSCLC is caught early and treated with surgery, the 5-year survival rate is around 65%. But, unfortunately, half of lung cancers are diagnosed at a distant stage, when the cancer has spread to the second lung or other organs3,4. These cancers are treated with radiation and systemic therapy such as chemotherapy, and the 5-year survival rate drops to less than 10%3.
Around 10% of NSCLC patients have more than one nodule (radiographically suspected tumor) at diagnosis5. (This number is rising due to increased use of chest CT scans, especially for high-risk individuals like smokers6.) There’s a big debate in the field about the causes, best ways to determine tumor stage, and best treatment for these multiple nodules. In part, these answers appear to depend on the details of the patient population.
In smokers, these nodules are often thought to be widespread but independent primary tumors caused by smoking-related DNA damage7. However, many studies have found overlapping mutations between nodules, suggesting a clonal relationship, which could indicate advanced disease and possible metastasis8-10.
Separately, about 15% of NSCLC cases are driven by EGFR mutations, which are more common in never-smokers, women, and Asian populations11-14. When these patients also have multiple nodules at diagnosis, clinicians are faced with unique questions. Patients with multiple early-stage EGFR-mutant tumors likely don’t have widespread carcinogen-induced DNA damage or late-stage disease — but then what’s driving their cancer, and what’s the best treatment course?
How we classify these nodules — either as independent tumors or advanced disease — affects treatment decisions and ultimately patient outcomes15.
This study aimed to investigate the mechanisms driving the growth of multiple early-stage EGFR-mutant lung tumors. We set out to answer two key questions:
- Are the multiple nodules in these patients independent primary tumors or metastases?
- What is the lineage relationship between multiple nodules in a single patient?
Understanding these questions is crucial for improving diagnosis, management, and surveillance strategies for patients with multiple EGFR-mutant lesions.
Methods
To explore the genetic origins of multiple early-stage EGFR-mutant lung cancers, we analyzed and reconstructed the phylogenetic relationships of tumors from patients with either familial or sporadic cases of multiple primary lung cancers. We included ten patients with multiple synchronous EGFR-mutant tumors, and members of a family with the germline EGFR T790M mutation, which predisposes them to multiple lung tumors16.
Our approach combined whole exome sequencing of tumor samples to identify driver and passenger mutations, with poly-G fingerprint analysis to measure genetic divergence (the evolutionary distance from the most recent common ancestor) between tumor samples17-21. We also used droplet digital PCR to detect ultra-rare mutations in normal lung tissues. These lineage tracing techniques allowed us to quantify the evolutionary relationships between tumors. The data provide a detailed view of the shared ancestry and genetic divergence among tumors within individual patients, allowing us to quantify not just if the tumors had a common ancestor, but approximately when they diverged. These results helped us infer the mechanisms of multiple tumor development in these patients.
Results
Our study identified four distinct mechanisms contributing to the emergence of multiple early-stage EGFR-mutant lung tumors.
Consistent with current models in the field:
- Independent Primary Tumors: One patient had tumors with no lineage relationship, indicating independent primary tumors. This patient had a light smoking history but quit decades before developing cancer. Air pollution may also play a role in EGFR-mutant cancer development, similar to smoking22.
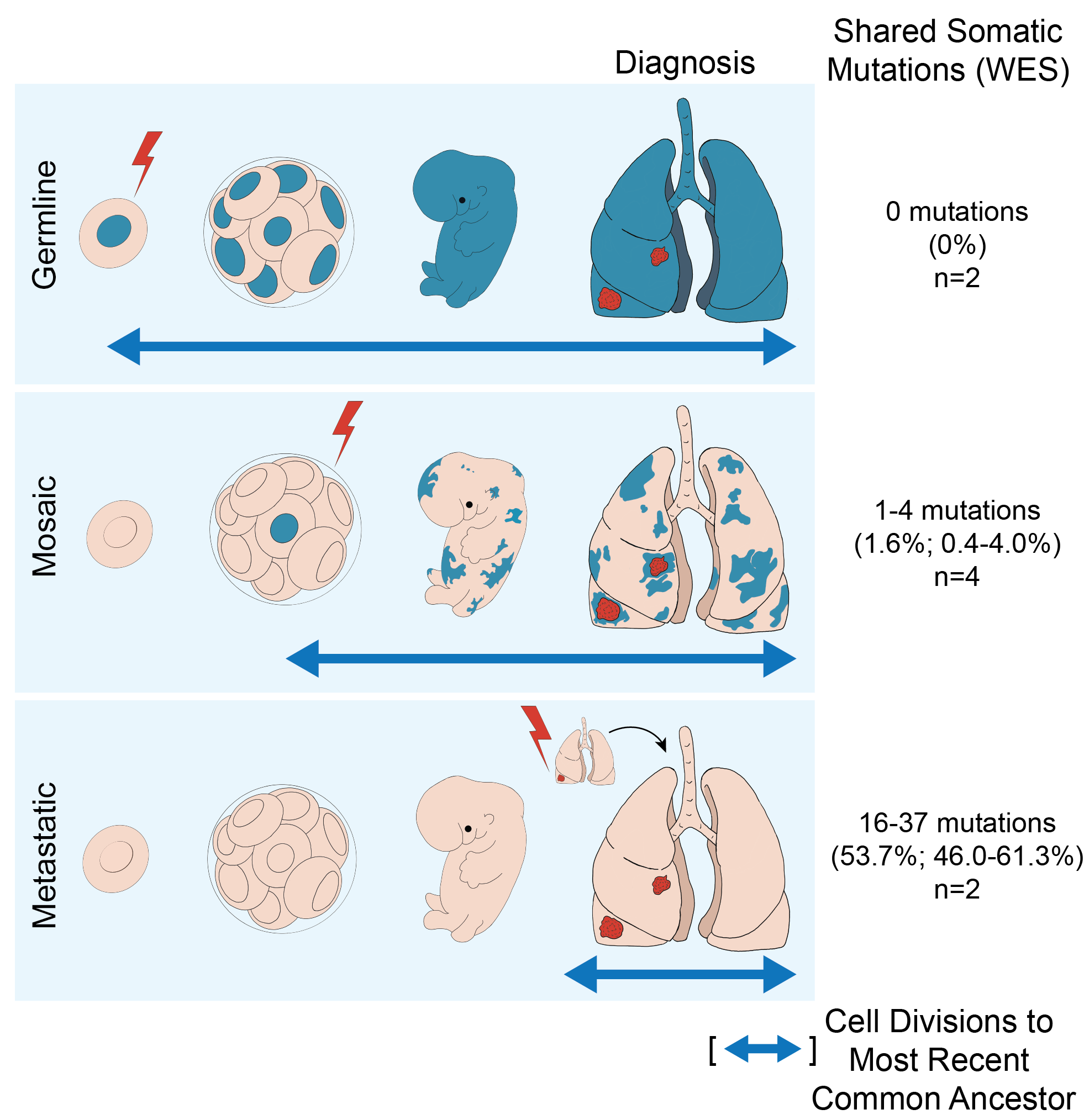
- Recurrence or Metastasis: Two patients had tumors with a clear, recent common ancestor. Their lineage trees showed long trunks and short branches, reflecting a high proportion of shared mutations before divergence. This pattern likely indicates recurrence or metastasis.
Unexpected mechanisms of genetic predisposition:
- Germline EGFR Mutation: Three patients had novel EGFR germline variants of unknown significance, such as G873E and H988P. Functional assays showed these variants have modest activating effects, suggesting they may promote tumorigenesis under certain conditions. Although inherited EGFR mutations are known to cause multiple tumor syndromes, this phenomenon was considered exceedingly rare. In the familial cases with the T790M EGFR mutation that we analyzed, multiple tumors arose independently from a germline mutated background, acquiring somatic canonical EGFR mutations in cis with the germline variant. These tumors shared no additional mutations, confirming the independent origins of these multiple primary tumors on top of the shared genetic background.
- Developmental Mosaicism for Mutant EGFR: In the remaining four sporadic cases, we found evidence of a new phenomenon called developmental mosaicism. EGFR mutations likely arose early during fetal lung development, creating a mosaic of cells throughout the lung tissue, some of which were predisposed to tumor development and others which were not. This was supported by the presence of a small number of shared somatic mutations among anatomically distinct tumors within individual patients. Their lineage trees had short trunks and long branches, indicating a small proportion of shared mutations. Poly-G fingerprint analysis confirmed a common evolutionary history that was intermediate between fully independent primary tumors and metastatic recurrences. We also observed a low variant allele fraction of EGFR-mutant DNA in the normal lung tissue of these patients, consistent with the presence of rare mosaic cells that had not undergone oncogenic conversion.
Implications
Our findings have significant implications for diagnosing and managing patients with multiple early-stage EGFR-mutant lung cancers. Traditionally, it’s been thought that these synchronous EGFR-mutant lung lesions are either independent primary tumors or the result of metastatic spread. But our study challenges this view. We propose that developmental mosaicism can lead to multiple primary tumors with shared genetic ancestry, a phenomenon that’s been largely overlooked in clinical settings. This concept isn’t just limited to lung cancer—it might apply to other cancer types as well.
This new understanding highlights the need for thorough genetic and histopathological evaluation of synchronous lung lesions. It’s crucial to distinguish between truly independent tumors, metastases, and mosaic-derived tumors because these distinctions may directly impact treatment decisions. For instance, should clinicians recommend curative surgery or opt for systemic therapies typically reserved for metastatic disease? And what kind of monitoring should a patient undergo after their initial treatment course?
Our discovery of germline variants contributing to EGFR-mutant lung cancer underscores the importance of genetic screening and surveillance in patients with multiple primary tumors, including those without known familial cancer syndromes. Patients identified with germline or mosaic EGFR mutations might benefit from enhanced monitoring to detect and manage new lesions early, potentially improving their outcomes. Most importantly, patients with mosaic-derived tumors that are histopathologically early stage should be considered for surgery even if their tumors share a handful of mutations because these mutations reflect developmental mosaicism and not advanced disease.
Overall, this study deepens our understanding of the genetic origins of multiple primary EGFR-mutant lung cancers and highlights the complexity of diagnosing and managing these cases. Future research should focus on identifying additional genetic variants that predispose individuals to multiple tumors, developing methods for detecting developmental mosaicism, and exploring potential preventive strategies for at-risk populations.
References:
- Bray, F., et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 74, 229-263 (2024).
- Society, A.C. Cancer Facts & Figures 2022. (Atlanta, 2022).
- Program, S.R. SEER*Explorer: An interactive website for SEER cancer statistics. (National Cancer Institute).
- Prevention, C.f.D.C.a. U.S. Cancer Statistics Lung Cancer Stat Bite. (U.S. Department of Health and Human Services, 2024).
- Detterbeck, F.C., et al. The IASLC Lung Cancer Staging Project: Summary of Proposals for Revisions of the Classification of Lung Cancers with Multiple Pulmonary Sites of Involvement in the Forthcoming Eighth Edition of the TNM Classification. J Thorac Oncol 11, 639-650 (2016).
- National Lung Screening Trial Research, T., et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365, 395-409 (2011).
- Gazdar, A.F. & Minna, J.D. Multifocal lung cancers--clonality vs field cancerization and does it matter? J Natl Cancer Inst 101, 541-543 (2009).
- Murphy, S.J., et al. Using Genomics to Differentiate Multiple Primaries From Metastatic Lung Cancer. J Thorac Oncol 14, 1567-1582 (2019).
- Wang, X., et al. Evidence for common clonal origin of multifocal lung cancers. J Natl Cancer Inst 101, 560-570 (2009).
- Schneider, F. & Dacic, S. Histopathologic and molecular approach to staging of multiple lung nodules. Transl Lung Cancer Res 6, 540-549 (2017).
- Skoulidis, F. & Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 19, 495-509 (2019).
- Jamal-Hanjani, M., et al. Tracking the evolution of non-small-cell lung cancer. New England Journal of Medicine 376, 2109-2121 (2017).
- Devarakonda, S., et al. Genomic Profiling of Lung Adenocarcinoma in Never-Smokers. J Clin Oncol 39, 3747-3758 (2021).
- Bell, D.W., et al. Increased prevalence of EGFR-mutant lung cancer in women and in East Asian populations: Analysis of estrogen-related polymorphisms. Clinical Cancer Research 14, 4079-4084 (2008).
- Leventakos, K., et al. Management of Multifocal Lung Cancer: Results of a Survey. J Thorac Oncol 12, 1398-1402 (2017).
- Bell, D.W., et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 37, 1315-1316 (2005).
- Naxerova, K., et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55-60 (2017).
- Naxerova, K., et al. Quantifying cell divisions along evolutionary lineages in cancer. (2024).
- Parikh, A.R., et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 25, 1415-1421 (2019).
- Costello, M., et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 41, e67 (2013).
- Leshchiner, I., et al. Inferring early genetic progression in cancers with unobtainable premalignant disease. Nat Cancer 4, 550-563 (2023).
- Hill, W., et al. Lung adenocarcinoma promotion by air pollutants. Nature 616, 159-167 (2023).
Follow the Topic
-
Nature Cancer

This journal aims to provide a unique forum through which the cancer community will learn about the latest, most significant cancer-related advances across the life, physical, applied and social sciences.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in