Unravelling links between neonatal sepsis outbreak strains and carbapenemase-carrying bacteria colonising hospital surfaces in low- and middle- income countries
Published in Microbiology
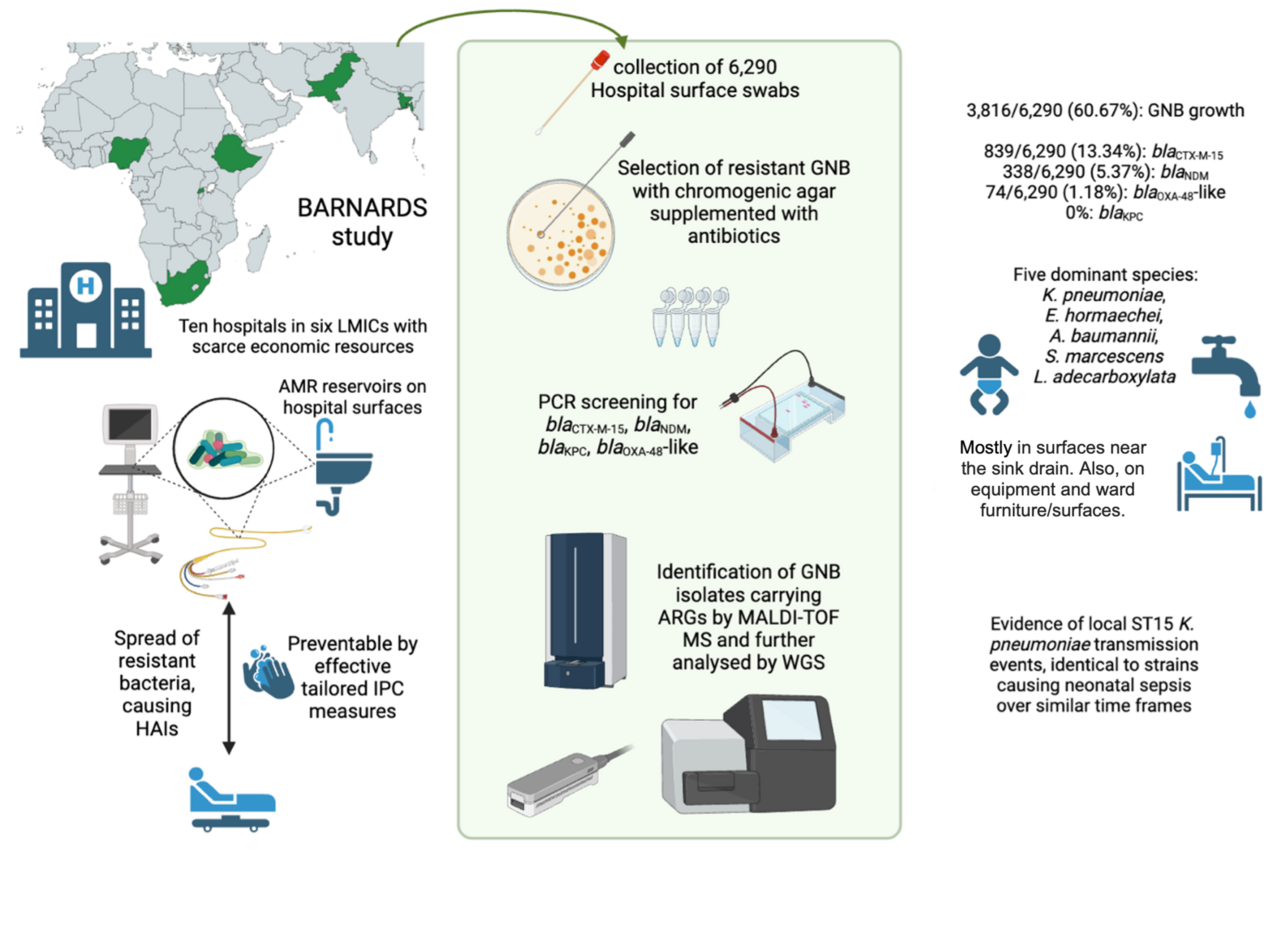
The BARNARDS study (Burden of Antibiotic Resistance in Neonates from Developing Societies, 2015-2018) determined the prevalence of antimicrobial resistance (AMR) bacteria in the gut microbiota of mothers and neonates, and characterised those isolates causing neonatal sepsis1,2. Isolates causing neonatal bloodstream infections showed resistance against clinically important antibiotics for neonatal sepsis treatment, as recommended by the world health organisation (WHO)3.
Carbapenem-resistant gram-negative bacteria (GNB) pose a significant public health threat in both high- (HIC) and low-and-middle-income (LMIC) countries. However, no evidence-based guidelines for prevention of these resistant bacteria in LMIC healthcare settings are accessible4. Further, the WHO and Médecins Sans Frontières still lack AMR data from many LMIC settings, thus global reports on infection prevention and control (IPC), burden of sepsis, or hospital acquired infections (HAIs) contain information mostly generated from HICs. Hospital surface colonisation by carbapenemase-carrying GNB is a significant concern in LMIC hospitals, where there may not be resources to implement appropriate IPC programs. Unless IPC practises are implemented, potential transmission to patients and thus HAI outbreaks may occur5.
In our group at University of Oxford and Cardiff University, we have been studying colonisation of patient care equipment and environmental surfaces by β-lactamase- and carbapenemase-producing bacteria, GNB diversity, and antimicrobial resistance genes (ARGs) across different hospital surface types from BARNARDS hospitals.
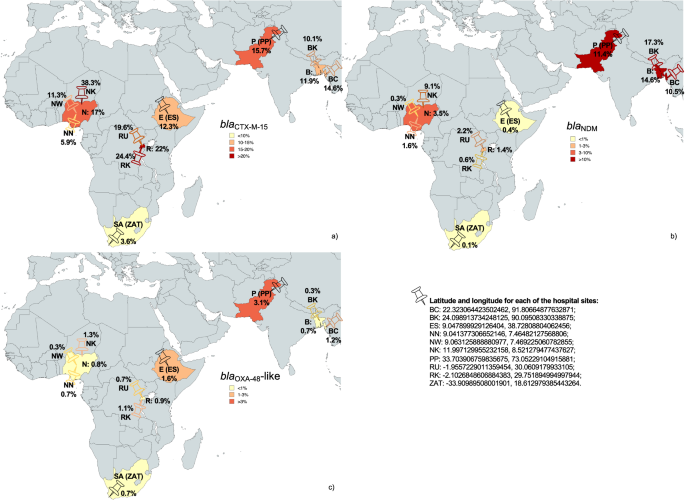
In this study, enrolling 10 hospital sites from 6 LMICs, we report a high prevalence of bacteria carrying ESBL (blaCTX-M-15), and carbapenemase genes (blaNDM and blaOXA-48-like) colonising surfaces near the sink drain. GNB carrying these genes were also recovered from medical equipment, ward furniture and surfaces, patients’ zones, and emergency neonatal care surfaces across all sites.
18 bacterial species were identified via whole genome sequencing, the most dominant being Klebsiella pneumoniae, Enterobacter hormaechei, Acinetobacter baumannii, Serratia marcescens and Leclercia adecarboxylata. High-risk double carbapenemase sequence type (ST)15 K. pneumoniae clone was recovered both from hospital surfaces and blood cultures from septic neonates in Pakistan over a 2-year period, suggesting this strain was colonising surfaces in the ward whilst simultaneously causing neonatal sepsis. These findings are alarming from an IPC point of view, suggesting the procedures in place are not effectively preventing HAIs in newborns.
Therefore, we investigated whether potential horizontal transmission of plasmids carrying ARGs occurred. We evidenced similarity between plasmids detected in ST15 K. pneumoniae and ST405 E. coli from the same hospital wards in Pakistan (IncA/C blaNDM-1 and the ColKP3-IncX3 blaOXA-181 plasmids), suggesting potential plasmid transmission across multiple hospital surfaces. We also studied genomic relatedness of clusters of the same strain of different GNB species (e.g., ST52 A. baumannii, ST231, ST316 and ST418 E. hormaechei, S. marcescens), which were detected from hospital surfaces in Pakistan, Bangladesh, and Rwanda, respectively. SNP analysis of ST52 A. baumannii revealed the sepsis isolate (identified in the same hospital in Rwanda) to be genetically distant (>100 SNPs) from the isolates collected from hospital surfaces, which were within 10 SNPs. Clusters of ST231 and ST316 (blaNDM-1 and mcr-9) E. hormaechei isolates recovered across surfaces in PP, a cluster of seven ST418 E. hormaechei from BK (recovered during a six-week period), were within less than five pairwise SNPs.
In this study we investigated correlation between colonisation and ARG prevalence, and time periods. β-lactamase and carbapenemase-carrying GNB were more frequently detected between March and October. Apart from IPC practises, other factors might be influencing bacterial reservoirs at certain times of the year, including climate, routine cleaning, and healthcare worker shifts. Further consideration should be taken to perform seasonal analyses at each site, as climate can be different even in different areas within the same country (e.g., South Africa). Our study holds particular importance, as it reports differences observed between countries but also hospital sites, suggesting IPC guidelines should be tailored at a hospital level. Support action plans should consider these findings, as well as stakeholders. Economic support would be needed to carry out more studies at this scale, to produce a large dataset to gain awareness of the current scenario.
This multinational scale study emphasises the global issue the bacterial contamination of hospital surfaces and the prevalence of AMR in these bacteria. Neonatal sepsis rates associated to AMR reservoirs on hospital surfaces may be reduced by improving IPC guidelines. However, more research should be conducted to understand correlations between surface material characteristics and cleaning/ disinfectants products applied for each of these materials at a hospital level in LMICs, across different bacterial species and strains, and with reference to plasmid transmission dynamics. We are incorporating these aspects during our next phase of the BARNARDS project.
References:
- Carvalho, M. J. et al. Antibiotic resistance genes in the gut microbiota of mothers and linked neonates with or without sepsis from low- and middle-income countries. Microbiol. 7, 1337–1347 (2022).
- Sands, K. et al. Characterization of antimicrobial-resistant Gramnegative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Microbiol. 6, 512–523 (2021).
- Thomson, K. M. et al. Effects of antibiotic resistance, drug target attainment, bacterial pathogenicity and virulence, and antibiotic access and affordability on outcomes in neonatal sepsis: an International Microbiology and Drug Evaluation Prospective substudy (BARNARDS). Lancet Infect. Dis. 21, 1677–1688 (2021).
- World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in Health Care Facilities. (Geneva, 2017).
- CDC and ICAN. Best Practices for Environmental Cleaning in Resource-Limited Healthcare Settings. A Healthcare Cleaning and Disinfection Guide for Healthcare Settings with Limited Resources (CDC and ICAN, 2023).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in