Unveiling TGFβ-SMAD signaling and DNA methylation in stem cells
Published in Cell & Molecular Biology

Every scientific paper is the result of countless hours of effort, collaboration, and discovery. Our recently published study in Nature Communications, A Stepwise Mode of TGFβ-SMAD Signaling and DNA Methylation Regulates Naïve-to-Primed Pluripotency and Differentiation (Nat Commun 15, 10123, 2024. https://doi.org/10.1038/s41467-024-54433-5), was no exception. Here's the story behind the scenes of this research journey.
A Long-Standing Mystery
For years, the canonical TGFβ signaling pathway has been a cornerstone in understanding embryogenesis. However, one enigma persisted: how Smad2/3 could function independently of Smad4 in specific contexts. The field needed a deeper dive into the intricacies of TGFβ signaling. This mystery captivated our curiosity. With our expertise in stem cell biology and molecular signaling, we set out to decipher this puzzle.
The Turning Point: Discovering Dnmt3b
A pivotal moment in this research came during our immunoprecipitation (IP) and mass spectrometry experiments. While exploring protein interactions in Smad4-knockout cells, we identified Dnmt3b—a key player in DNA methylation—as a surprising partner of Smad2/3. This discovery opened new avenues of exploration. Could Dnmt3b act as a mediator for Smad2/3 activity independently of Smad4?
The finding was both thrilling and daunting. We knew this would require rigorous validation across multiple models and techniques, from co-IP, pull-down to ChIP-seq and CUT&Tag, to confirm Dnmt3b's role.
Challenges Along the Way
Science rarely follows a straight path, and our journey was no different. One of the biggest challenges was distinguishing the distinct roles of Smad2 and Smad3 versus Smad4. This required creating and analyzing multiple knockout models (Smad2/3 double knockout, Smad4 single knockout, and even Smad2/3/4 triple knockout). Each model revealed nuanced differences in signaling and gene regulation, highlighting the complexity of these pathways.
Another hurdle was understanding how Dnmt3b facilitates Smad2/3 function. This required integrating epigenomics data, such as DNA methylation profiles, histone modification patterns, genome-wide binding signatures of Dnmt3b and Smad2/3, with transcriptomics data. The bioinformatics analysis was intricate and time-intensive but ultimately rewarding.
A Stepwise Paradigm Emerges
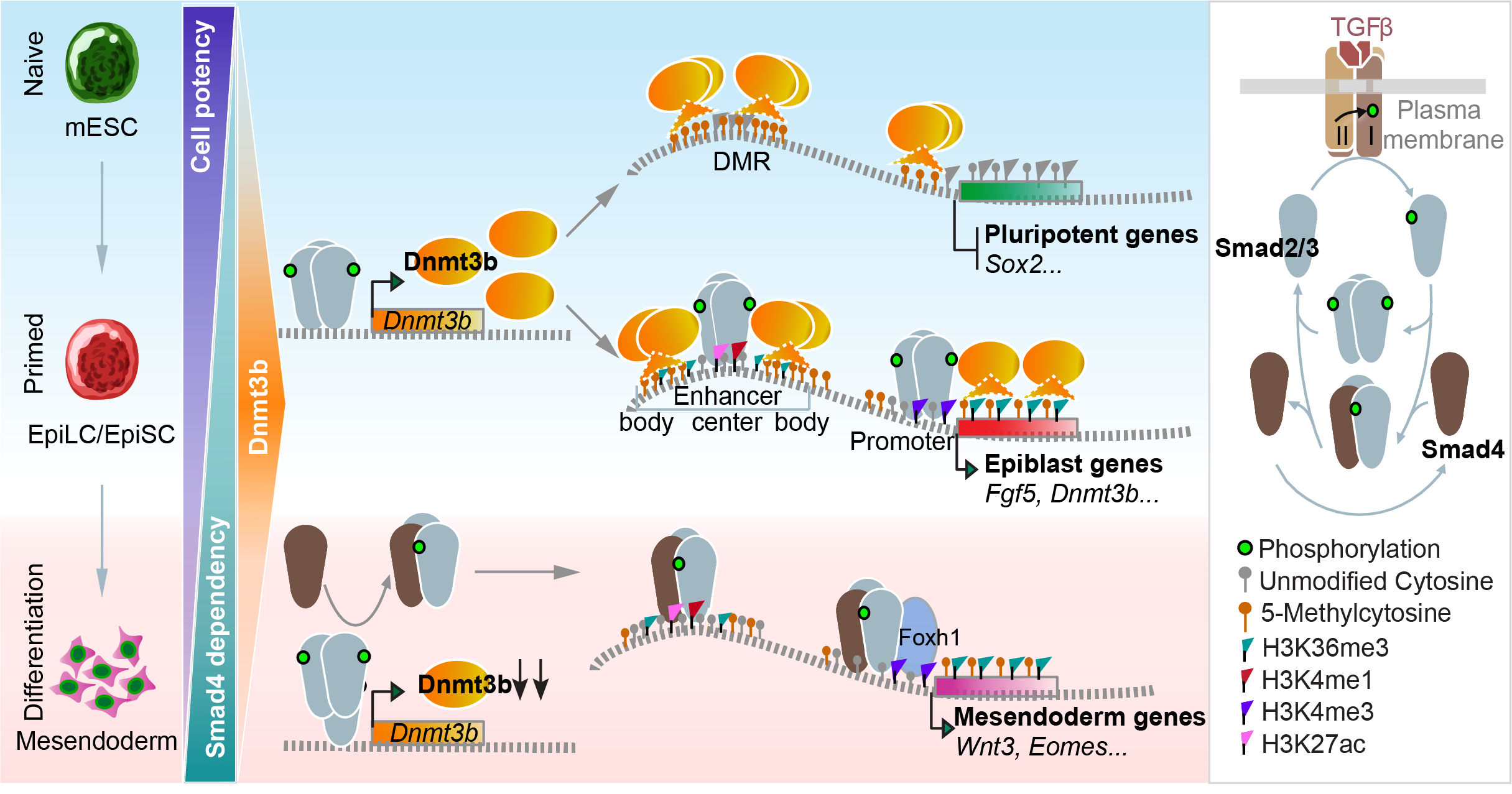
A two-step model describes the transition from naïve to primed epiblast and subsequent differentiation.
Through this work, we uncovered a stepwise regulatory mechanism:
- Naïve-to-Primed Transition: Smad2/3 upregulates Dnmt3b independently of Smad4, which establishes proper DNA methylation patterns, and enables Smad2/3 binding to the epiblast marker genes.
- Mesendoderm Differentiation: Dnmt3b becomes less actively engaged in global genome methylation, and Smad4 steps in to collaborate with Smad2/3, forming the canonical Smad complex necessary for mesendoderm lineage commitment.
Our two-step model is also consistent with the in vivo data. Conditional knockout of Smad4 in the epiblast using the Cre-LoxP system does not prevent the development from the epiblast to the gastrulation initiation, but shows focal defects in the primitive streak. As gastrulation proceeds, Smad4-deficient embryos fail to form derivatives of the anterior primitive streak, including definitive endoderm, sharing many phenotypic similarities with the downregulation of the Nodal/Smad2/3/Foxh1 pathway. This relay-like mechanism provided a new framework for understanding how TGFβ signaling orchestrates early embryonic development.
Sharing Our Findings with the Scientific Community
Science thrives on curiosity and the willingness to question assumptions. This work exemplifies how revisiting foundational concepts can lead to unexpected discoveries. Our study not only sheds light on fundamental developmental processes but also has broader implications. Understanding how TGFβ signaling and DNA methylation intersect could inform research into regenerative medicine and cancer biology, where these pathways play critical roles.
Feel free to dive into the details of our study here, and don’t hesitate to reach out with questions or thoughts. Science is, after all, a collective journey!

Article link:
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in