Unveiling the Mystery of Fungal RNA Editing Machinery: A Story of Unexpected Discoveries
Published in Microbiology, Cell & Molecular Biology, and Plant Science

Our journey began with a well-established fact: RNA editing, the alteration of RNA molecules after they've been copied from DNA, can be a powerful tool for gene therapy1. But within this world of editing, one type stood out - A-to-I mRNA editing. This process modifies a vast number of RNA instructions (mRNAs) made from our genes. It was first discovered in frogs 2, and for decades, we thought animal enzymes (ADAR) were the only players. Then, in 2016, our team stumbled upon something unexpected - A-to-I mRNA editing in a plant pathogenic fungus, Fusarium graminearum 3. This challenged everything we thought we knew, because ADAR genes are exclusive to animals!
Intrigued, we delved into the fungal genome, searching for the culprit. Our meticulous detective work led us to two genes, TAD2 and TAD3 4. These were already known to work together in eukaryotes, but for a different kind of RNA editing - on tRNA molecules 5, 6, 7. This finding aligned with what we knew about bacteria, where TadA, a homolog of the Tad2-Tad3 complex, mediated A-to-I mRNA editing 8, 9.
But a key question remained: how does this tRNA-editing complex recognize and edit mRNA, a completely different molecule? Additionally, this editing in fungi only happened during sexual reproduction 10. It seemed the puzzle had even more pieces!
Sometimes, the simplest approaches lead to breakthroughs. We initially tried to isolate the interacting proteins directly, but this hit a dead end. So, we shifted gears and focused on genes specifically expressed during fungal sexual reproduction. This time-consuming but targeted strategy led us to a crucial gene we renamed Ame1. Our combined evidence revealed a new editing complex: Tad2-Tad3-Ame1 (Fig.1). This wasn't just about understanding fungi; it shed light on how RNA editing systems might have evolved across all living things.
The next step was figuring out how Ame1 empowers the Tad2-Tad3 complex. Normally, this complex recognizes tRNA based on its 3D structure 5, 6, 7. Our research showed that Ame1 interacts with Tad3, acting like a key that unlocks the complex's ability to recognize mRNA as well. This discovery explains how a tRNA-editing complex evolved to edit mRNA – Ame1 acts as a cofactor, expanding the complex's editing capabilities.
But why does this editing only happen during fungal sexual reproduction? There are several reasons (Fig.1). First, Ame1 is only expressed then. Second, Tad2 and Tad3 undergo changes during this stage, becoming more efficient editors. Finally, Tad3 itself gets an extra boost through modifications. This stage-specific editing is driven by natural selection. Our research showed that editing benefits fungal sexual reproduction, but hinders their survival. It suggests that fungi have evolved this editing system to optimize their life cycle, increasing both reproductive success and survival strategies. These findings align with our previous work on the adaptive advantages conferred by A-to-I editing sites in fungi 11, 12, 13.
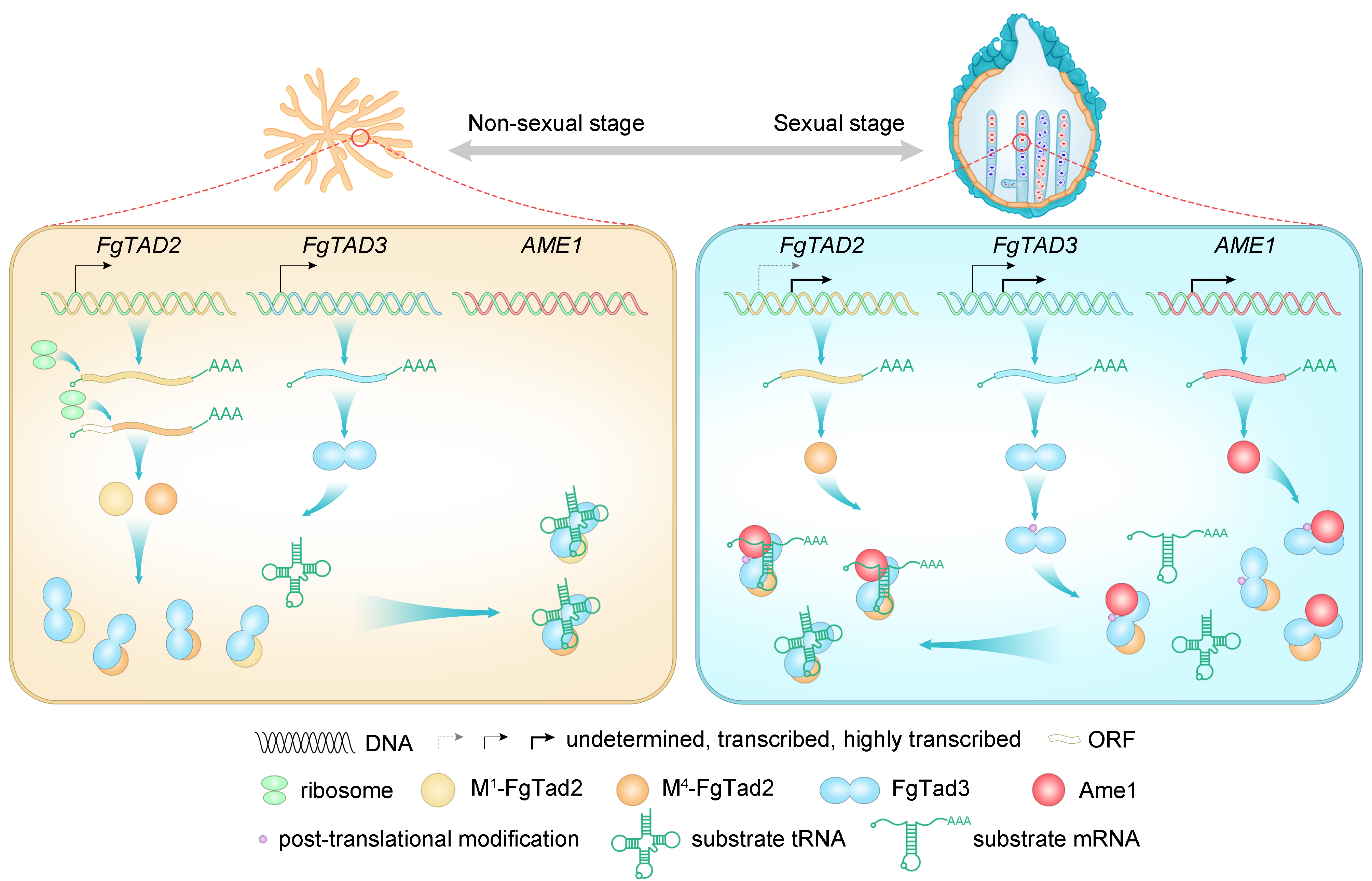
Finally, our findings suggest a way to predict which fungi can edit mRNA. We discovered that Ame1, although widespread, underwent a key duplication event in the ancestor of Sordariomycetes fungi. This led to a modified Ame1 that could interact with the editing complex, enabling A-to-I mRNA editing in this specific fungal lineage. By studying these changes, we might be able to predict this editing capability in other fungi.
This research goes far beyond understanding fungal biology. The Tad2-Tad3-Ame1 complex works in yeast, bacteria, plants, and human cell lines. This opens doors for exciting applications:
- Gene Editing Tools: This system could be harnessed to develop new tools for fixing faulty genes in humans (gene therapy) or improving crops (agriculture).
- Controlling Fungal Diseases: Targeting this editing system could be a new way to combat fungal plant diseases.
- Designing New Enzymes: Understanding how Ame1 interacts with other enzymes might inspire the creation of entirely new enzymes with unique abilities.
Unraveling the fungal A-to-I mRNA editing system has been a fascinating journey. It's a story of unexpected discoveries, the power of adaptation, and the potential for this knowledge to advance our understanding of life and its applications.
References:
- Khosravi HM, Jantsch MF. Site-directed RNA editing: recent advances and open challenges. RNA biology 18, 41-50 (2021).
- Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089-1098 (1988).
- Liu H, et al. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res 26, 499-509 (2016).
- Sun ML, et al. Stage-specific regulation of purine metabolism during infectious growth and sexual reproduction in Fusarium graminearum. New Phytologist 230, 757-773 (2021).
- Liu X, et al. Crystal structure of the yeast heterodimeric ADAT2/3 deaminase. Bmc Biol 18, 189 (2020).
- Ramos-Morales E, et al. The structure of the mouse ADAT2/ADAT3 complex reveals the molecular basis for mammalian tRNA wobble adenosine-to-inosine deamination. Nucleic Acids Res 49, 6529-6548 (2021).
- Dolce LG, et al. Structural basis for sequence-independent substrate selection by eukaryotic wobble base tRNA deaminase ADAT2/3. Nat Commun 13, 6737 (2022).
- Bar-Yaacov D, et al. RNA editing in bacteria recodes multiple proteins and regulates an evolutionarily conserved toxin-antitoxin system. Genome Research 27, 1696-1703 (2017).
- Liao WX, Nie WH, Ahmad I, Chen GY, Zhu B. The occurrence, characteristics, and adaptation of A-to-I RNA editing in bacteria: A review. Frontiers in microbiology 14, (2023).
- Bian Z, Ni Y, Xu JR, Liu H. A-to-I mRNA editing in fungi: occurrence, function, and evolution. Cell Mol Life Sci 76, 329-340 (2019).
- Qi Z, et al. Adaptive advantages of restorative RNA editing in fungi for resolving survival-reproduction trade-offs. Sci Adv 10, eadk6130 (2024).
- Xin K, et al. Experimental evidence for the functional importance and adaptive advantage of A-to-I RNA editing in fungi. Proc Natl Acad Sci U S A 120, e2219029120 (2023).
- Liu H, et al. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc Natl Acad Sci U S A 114, E7756-E7765 (2017).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in